3. How to run tests¶
For more example visit: https://github.com/KaliLab/HippoUnit_demo/
3.1. Run validation tests of HippoUnit on the Migliore et al. 2011 model of hippocampal CA1 pyramidal cell¶
The purpose of this notebook is to show how to run the validation tests of HippoUnit (https://github.com/KaliLab/hippounit) on the Migliore et al. 2011 CA1 pyramidal cell model (ModelDB accession number: 138205).
The tests shall take as input a standalone version of the model (a hoc file) that should not display any GUI, or contain any built in simulations and run-time modifications. The hoc file that is used in this notebook is available here: https://github.com/KaliLab/HippoUnit_demo/blob/master/published_models/Migliore_et_al_2011_Schizophr/main_model.hoc
Target experimental data that are used by the validation tests in this notebook are available here: https://github.com/KaliLab/HippoUnit_demo/tree/master/target_features
[1]:
from __future__ import print_function
%matplotlib inline
from hippounit.utils import ModelLoader
from hippounit import tests
from json2html import *
import pkg_resources
import json
import collections
import numpy
import IPython
3.2. Instantiating the model¶
The ModelLoader class of HippoUnit is instantiated and the model, and simulation related parameters are set.
[2]:
# path to mod files
mod_files_path = "../published_models/Migliore_et_al_2011_Schizophr/"
#all the outputs will be saved here. It will be an argument to the test.
base_directory = '../published_models_validation_results/'
#Load cell model
model = ModelLoader(mod_files_path = mod_files_path )
# outputs will be saved in subfolders named like this:
model.name="Migliore_et_al_2011"
# path to hoc file
# the model must not display any GUI!!
model.hocpath = "../published_models/Migliore_et_al_2011_Schizophr/main_model.hoc"
# If the hoc file doesn't contain a template, this must be None (the default value is None)
model.template_name = None
# model.SomaSecList_name should be None, if there is no Section List in the model for the soma, or if the name of the soma section is given by setting model.soma (the default value is None)
model.SomaSecList_name = None
# if the soma is not in a section list or to use a specific somatic section, add its name here:
model.soma = 'soma[0]'
# For the PSP Attenuation Test, and Back-propagating AP Test a section list containing the trunk sections is needed
model.TrunkSecList_name = 'trunk_sec_list'
# For the Oblique Integration Test a section list containing the oblique dendritic sections is needed
model.ObliqueSecList_name = 'oblique_dendrites'
#This will be argument to those tests, where dendritic locatins are selected according to distances. If not set, the end of the above given soma section will be used as reference point for distance determination
trunk_origin = ['soma[1]', 1]
# It is important to set the v_init and the celsius parameters of the simulations here,
# as if they are only set in the model's files, they will be overwritten with the default values of the ModelLoader class.
# default values: v_init = -70, celsius = 34
model.v_init = -65
model.celsius = 35
3.3. Instantiating and running the tests of HippoUnit on the model¶
At the end of each test, the user is provided with a textual summary of the score and the path to related output files generated by the tests.
3.3.1. Somatic Features Test - UCL data set¶
It evaluates the model against various eFEL (https://github.com/BlueBrain/eFEL) features under somatic current injection of varying amplitudes. The experimental dataset used for validation is obtained from sharp electrode recordings in rats at UCL (Migliore et al. 2018 https://doi.org/10.1371/journal.pcbi.1006423).
[3]:
# Load target data
with open('../target_features/feat_CA1_pyr_cACpyr_more_features.json') as f:
observation = json.load(f, object_pairs_hook=collections.OrderedDict)
[4]:
# Load stimuli file
ttype = "CA1_pyr_cACpyr"
stim_file = pkg_resources.resource_filename("hippounit", "tests/stimuli/somafeat_stim/stim_" + ttype + ".json")
with open(stim_file, 'r') as f:
config = json.load(f, object_pairs_hook=collections.OrderedDict)
# Instantiate test class
test = tests.SomaticFeaturesTest(observation=observation, config=config, force_run=False, show_plot=True, save_all = True, base_directory=base_directory)
# test.specify_data_set is added to the name of the subdirectory (somaticfeat), so test runs using different data sets can be saved into different directories
test.specify_data_set = 'UCL_data'
# Number of parallel processes
test.npool = 10
try:
#Run the test
score = test.judge(model)
#Summarize and print the score achieved by the model on the test using SciUnit's summarize function
score.summarize()
except Exception as e:
print('Model: ' + model.name + ' could not be run')
print(e)
pass
- running amplitude: -0.8 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: -1.0 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: -0.4 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
Changed dt
Changed dt
- running amplitude: -0.6 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.4 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: -0.2 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
Changed dt
Changed dt
- running amplitude: 0.2 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.6 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
Changed dt
- running amplitude: 1.0 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.8 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
Changed dt
The figures are saved in the directory: ../published_models_validation_results/figs/somaticfeat_UCL_data/Migliore_et_al_2011/
Number of features succesfully evaluated: 232/232
Results are saved in the directory: ../published_models_validation_results/results/somaticfeat_UCL_data/Migliore_et_al_2011/
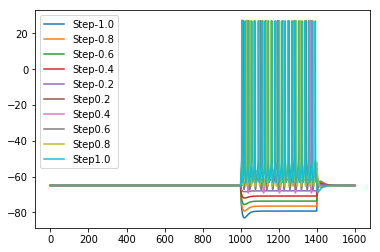
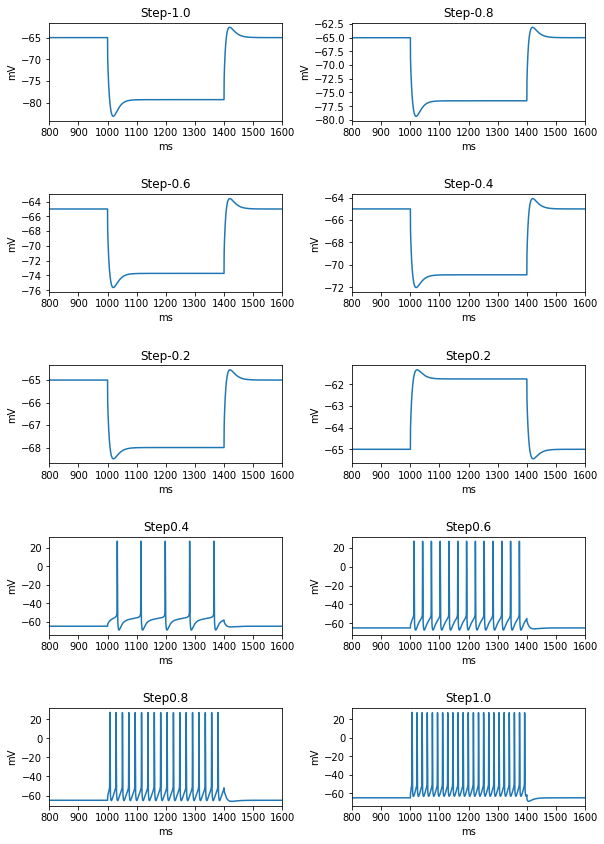
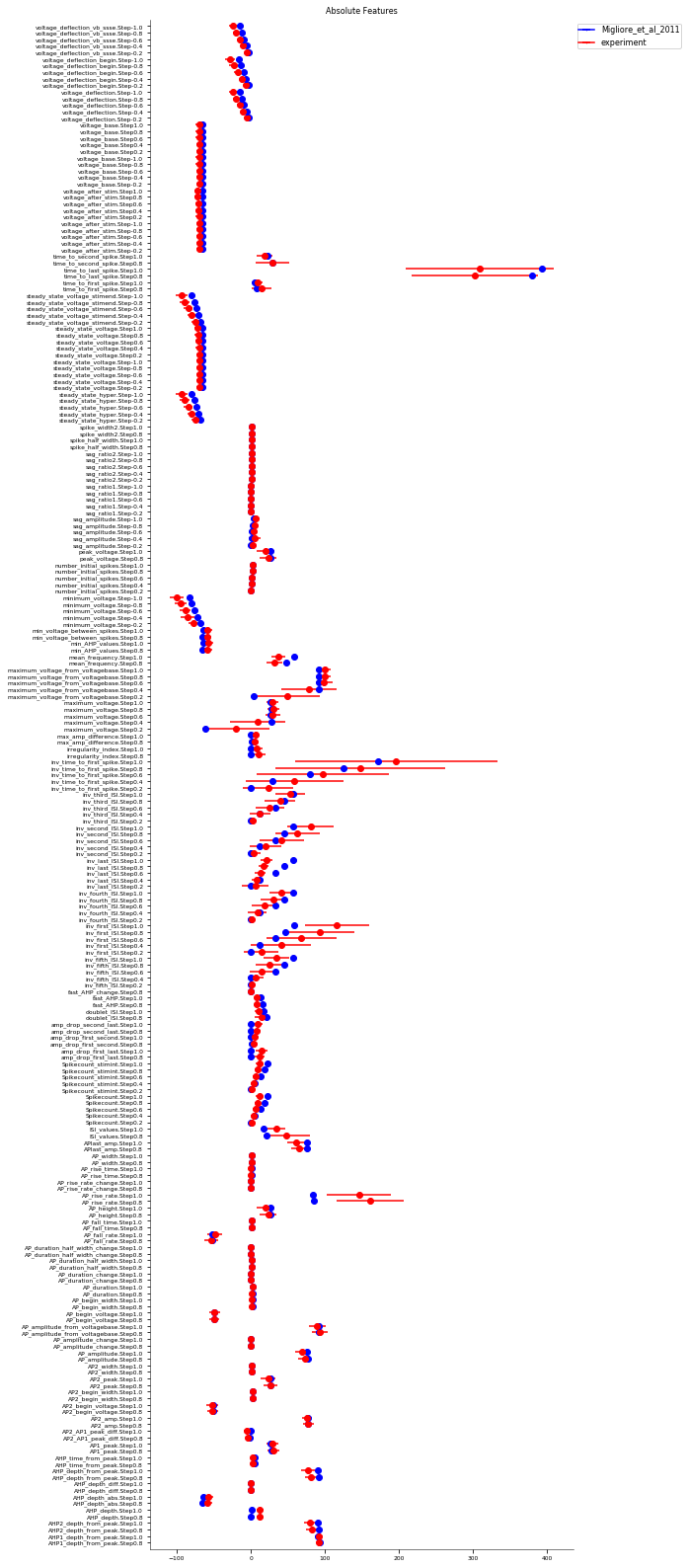
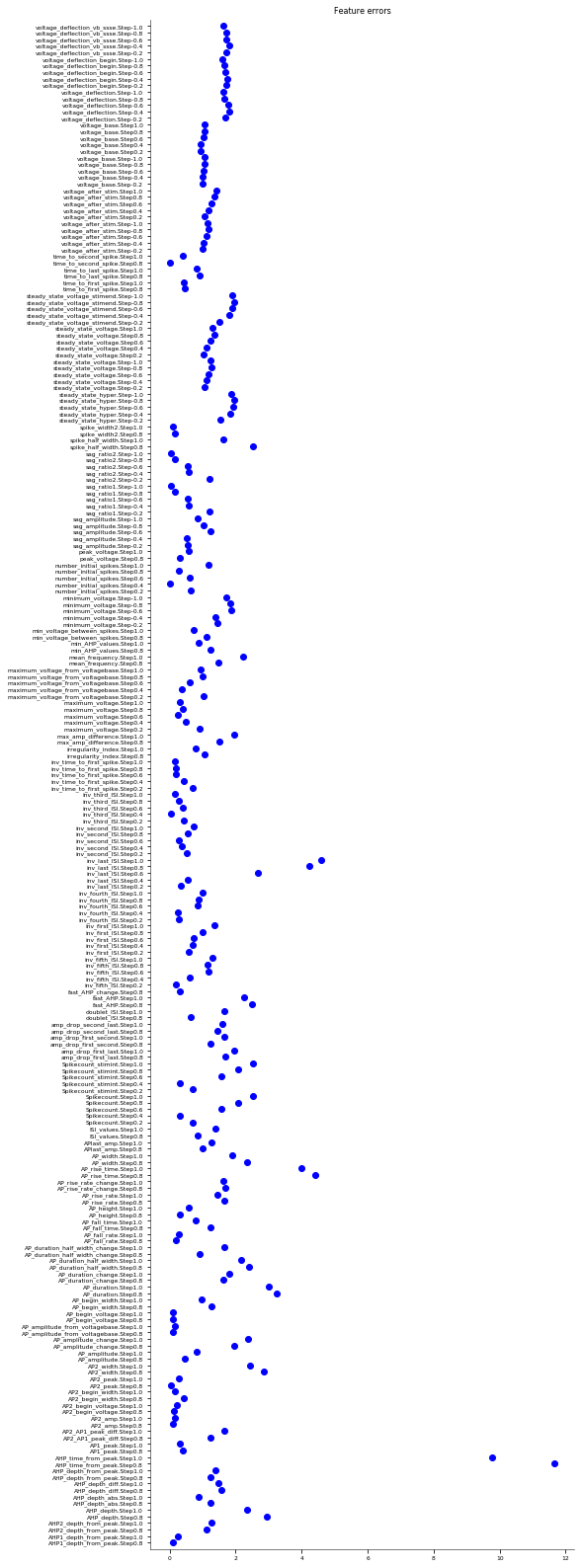
3.3.2. Somatic Features Test - JMakara data set¶
It evaluates the model against various eFEL (https://github.com/BlueBrain/eFEL) features under somatic current injection of varying amplitudes. The experimental dataset used for validation is obtained from patch clamp recordings in rats by Judit Makara
[5]:
# Load target data
with open('../target_features/feat_rat_CA1_JMakara_more_features.json') as f:
config_pyr = json.load(f, object_pairs_hook=collections.OrderedDict)
observation = config_pyr
[6]:
# Load stimuli file
stim_file = pkg_resources.resource_filename("hippounit", "tests/stimuli/somafeat_stim/stim_rat_CA1_PC_JMakara.json")
with open(stim_file, 'r') as f:
config = json.load(f, object_pairs_hook=collections.OrderedDict)
# Instantiate test class
test = tests.SomaticFeaturesTest(observation=observation, config=config, force_run=False, show_plot=True, save_all = True, base_directory=base_directory)
# test.specify_data_set is added to the name of the subdirectory (somaticfeat), so test runs using different data sets can be saved into different directories
test.specify_data_set = 'JMakara_data'
# Number of parallel processes
test.npool = 10
try:
#Run the test
score = test.judge(model)
#Summarize and print the score achieved by the model on the test using SciUnit's summarize function
score.summarize()
except Exception as e:
print('Model: ' + model.name + ' could not be run')
print(e)
pass
- running amplitude: -0.1 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: -0.25 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: -0.2 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: -0.15 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.05 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.1 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: -0.05 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
- running amplitude: 0.15 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
Changed dt
- running amplitude: 0.2 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.25 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
Changed dt
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AHP_depth:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AHP_depth:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AHP_depth:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AHP_depth_abs:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AHP_depth_abs:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AHP_depth_abs:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AHP_depth_diff:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_values] is missing
Feature [AHP_depth] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AHP_depth_diff:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_values] is missing
Feature [AHP_depth] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AHP_depth_diff:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_values] is missing
Feature [AHP_depth] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AHP_depth_from_peak:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AHP_depth_from_peak:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AHP_depth_from_peak:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AHP_time_from_peak:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AHP_time_from_peak:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AHP_time_from_peak:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_amplitude:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [peak_voltage] is missing
AP_amplitude: Error calculating peak_voltage
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_amplitude:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [peak_voltage] is missing
AP_amplitude: Error calculating peak_voltage
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_amplitude:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [peak_voltage] is missing
AP_amplitude: Error calculating peak_voltage
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_amplitude_change:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [peak_voltage] is missing
AP_amplitude: Error calculating peak_voltage
Feature [AP_amplitude] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_amplitude_change:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [peak_voltage] is missing
AP_amplitude: Error calculating peak_voltage
Feature [AP_amplitude] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_amplitude_change:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [peak_voltage] is missing
AP_amplitude: Error calculating peak_voltage
Feature [AP_amplitude] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_amplitude_from_voltagebase:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
Error calculating peak_voltage for AP_amplitude_from_voltagebase
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_amplitude_from_voltagebase:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
Error calculating peak_voltage for AP_amplitude_from_voltagebase
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_amplitude_from_voltagebase:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
Error calculating peak_voltage for AP_amplitude_from_voltagebase
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_begin_voltage:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_begin_voltage:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_begin_voltage:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_begin_width:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [min_AHP_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_begin_width:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [min_AHP_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_begin_width:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [min_AHP_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_duration:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_duration:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_duration:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_duration_change:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_duration] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_duration_change:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_duration] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_duration_change:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_duration] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_duration_half_width:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_rise_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_duration_half_width:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_rise_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_duration_half_width:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_rise_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_duration_half_width_change:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_rise_indices] is missing
Feature [AP_duration_half_width] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_duration_half_width_change:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_rise_indices] is missing
Feature [AP_duration_half_width] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_duration_half_width_change:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_rise_indices] is missing
Feature [AP_duration_half_width] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_height:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_height:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_height:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_rise_rate:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_rise_rate:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_rise_rate:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_rise_rate_change:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_rise_rate] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_rise_rate_change:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_rise_rate] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_rise_rate_change:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
Feature [AP_rise_rate] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_rise_time:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_rise_time:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_rise_time:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_width:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
No spike in trace.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_width:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
No spike in trace.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature AP_width:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
No spike in trace.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature ISI_CV:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
Feature [ISI_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature ISI_CV:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
Feature [ISI_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature ISI_log_slope:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
Feature [ISI_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature ISI_log_slope:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
Feature [ISI_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature ISI_log_slope_skip:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
Feature [ISI_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature ISI_log_slope_skip:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
Feature [ISI_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature ISI_semilog_slope:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
Feature [ISI_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature ISI_semilog_slope:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
Feature [ISI_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature ISI_values:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature ISI_values:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature ISI_values:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature adaptation_index:
Voltage never goes below or above threshold in spike detection.
Parameter [offset] is missing in double map. In the python interface this can be set using the setDoubleSetting() function
Minimum 4 spike needed for feature [adaptation_index].
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature adaptation_index:
Voltage never goes below or above threshold in spike detection.
Parameter [offset] is missing in double map. In the python interface this can be set using the setDoubleSetting() function
Minimum 4 spike needed for feature [adaptation_index].
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature adaptation_index2:
Voltage never goes below or above threshold in spike detection.
At least 4 spikes needed for adaptation_index2.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature adaptation_index2:
Voltage never goes below or above threshold in spike detection.
At least 4 spikes needed for adaptation_index2.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature amp_drop_first_last:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
At least 2 spikes needed for calculation of amp_drop_first_last.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature amp_drop_first_last:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
At least 2 spikes needed for calculation of amp_drop_first_last.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature amp_drop_first_last:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
At least 2 spikes needed for calculation of amp_drop_first_last.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature amp_drop_first_second:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
At least 2 spikes needed for calculation of amp_drop_first_second.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature amp_drop_first_second:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
At least 2 spikes needed for calculation of amp_drop_first_second.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature amp_drop_first_second:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
At least 2 spikes needed for calculation of amp_drop_first_second.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature amp_drop_second_last:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
At least 3 spikes needed for calculation of amp_drop_second_last.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature amp_drop_second_last:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
At least 3 spikes needed for calculation of amp_drop_second_last.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature amp_drop_second_last:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
At least 3 spikes needed for calculation of amp_drop_second_last.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature burst_mean_freq:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
Error: More than 5 spike is needed for burst calculation.
Feature [burst_ISI_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature burst_mean_freq:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
Error: More than 5 spike is needed for burst calculation.
Feature [burst_ISI_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature burst_number:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
Error: More than 5 spike is needed for burst calculation.
Feature [burst_ISI_indices] is missing
Feature [burst_mean_freq] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature doublet_ISI:
Voltage never goes below or above threshold in spike detection.
Need at least two spikes for doublet_ISI.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature doublet_ISI:
Voltage never goes below or above threshold in spike detection.
Need at least two spikes for doublet_ISI.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature doublet_ISI:
Voltage never goes below or above threshold in spike detection.
Need at least two spikes for doublet_ISI.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature fast_AHP:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature fast_AHP:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature fast_AHP:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature fast_AHP_change:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
Feature [fast_AHP] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature fast_AHP_change:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
Feature [AP_begin_indices] is missing
Feature [fast_AHP] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature irregularity_index:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
Feature [ISI_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature irregularity_index:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
Feature [ISI_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature irregularity_index:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
Feature [ISI_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature max_amp_difference:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
At least 2 spikes needed for calculation of max_amp_difference.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature max_amp_difference:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
At least 2 spikes needed for calculation of max_amp_difference.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature max_amp_difference:
Voltage never goes below or above threshold in spike detection.
Feature [peak_voltage] is missing
At least 2 spikes needed for calculation of max_amp_difference.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature mean_frequency:
Voltage never goes below or above threshold in spike detection.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature mean_frequency:
Voltage never goes below or above threshold in spike detection.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature mean_frequency:
Voltage never goes below or above threshold in spike detection.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature min_AHP_values:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature min_AHP_values:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature min_AHP_values:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature peak_voltage:
Voltage never goes below or above threshold in spike detection.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature peak_voltage:
Voltage never goes below or above threshold in spike detection.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature peak_voltage:
Voltage never goes below or above threshold in spike detection.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature single_burst_ratio:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
Feature [ISI_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature single_burst_ratio:
Voltage never goes below or above threshold in spike detection.
Three spikes required for calculation of ISI_values.
Feature [ISI_values] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature spike_half_width:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature spike_half_width:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature spike_half_width:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature spike_width2:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature spike_width2:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature spike_width2:
Voltage never goes below or above threshold in spike detection.
At least one spike required for calculation of min_AHP_indices.
Feature [min_AHP_indices] is missing
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature time_to_first_spike:
Voltage never goes below or above threshold in spike detection.
One spike required for time_to_first_spike.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature time_to_first_spike:
Voltage never goes below or above threshold in spike detection.
One spike required for time_to_first_spike.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature time_to_first_spike:
Voltage never goes below or above threshold in spike detection.
One spike required for time_to_first_spike.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature time_to_second_spike:
Voltage never goes below or above threshold in spike detection.
Two spikes required for time_to_second_spike.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature time_to_second_spike:
Voltage never goes below or above threshold in spike detection.
Two spikes required for time_to_second_spike.
RuntimeWarning)
/home/saray/venv_Python3/lib/python3.5/site-packages/efel/api.py:480: RuntimeWarning: Error while calculating feature time_to_second_spike:
Voltage never goes below or above threshold in spike detection.
Two spikes required for time_to_second_spike.
RuntimeWarning)
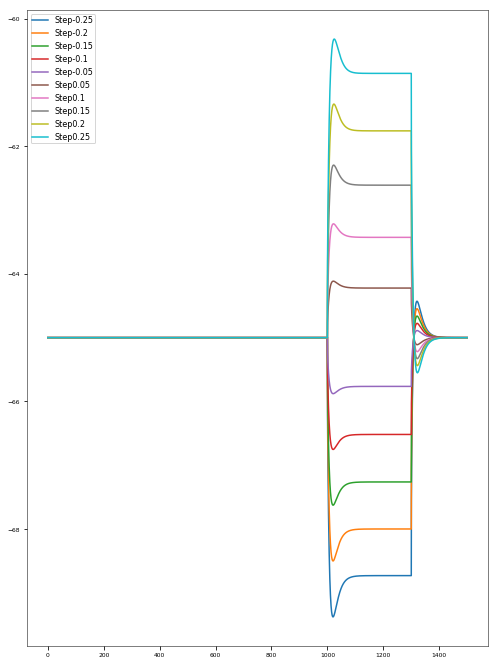
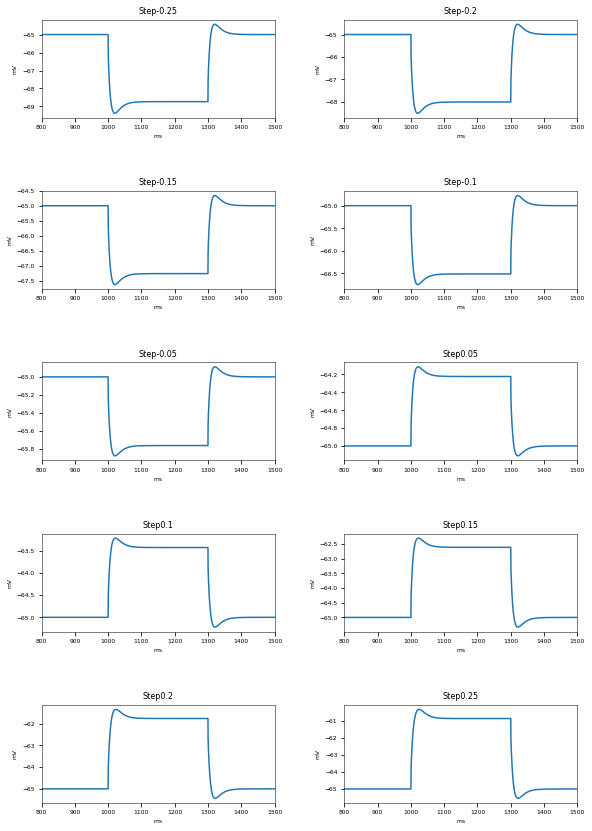
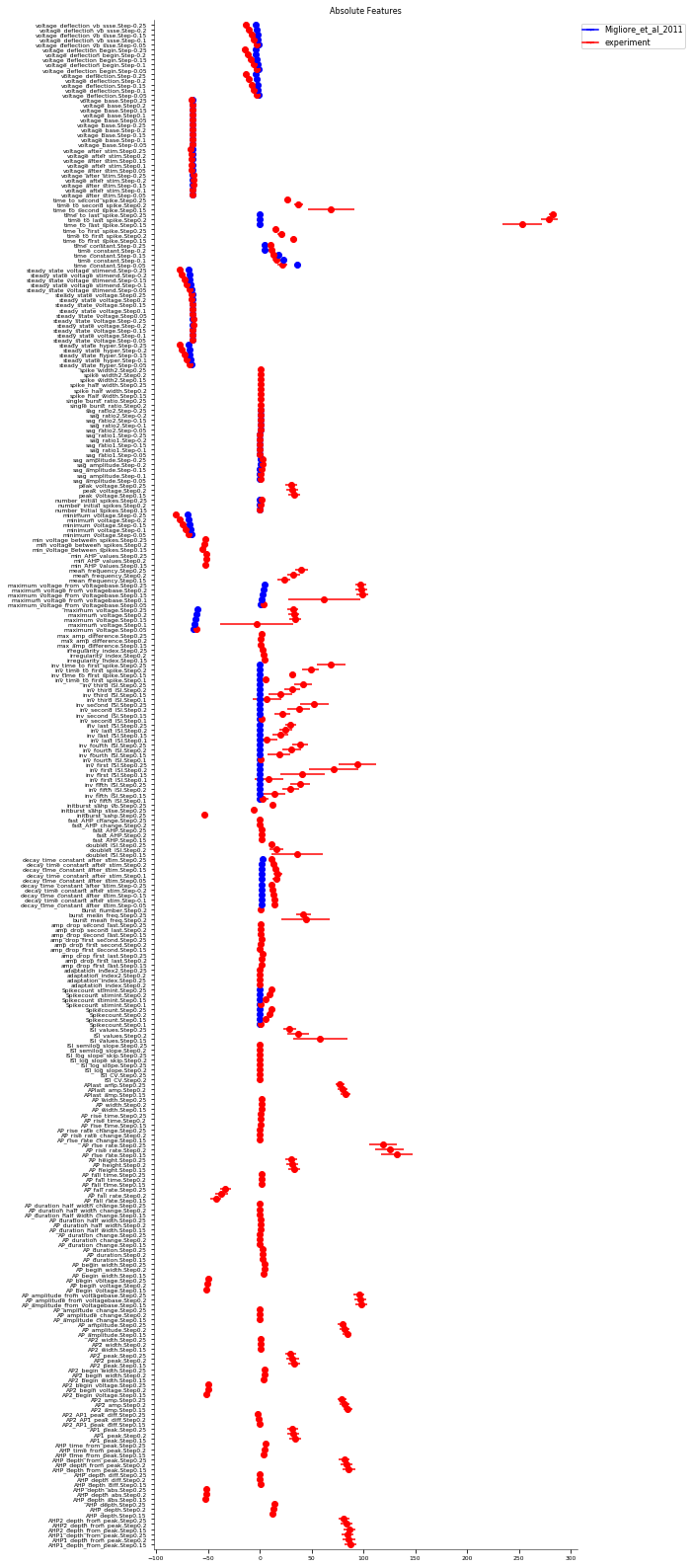
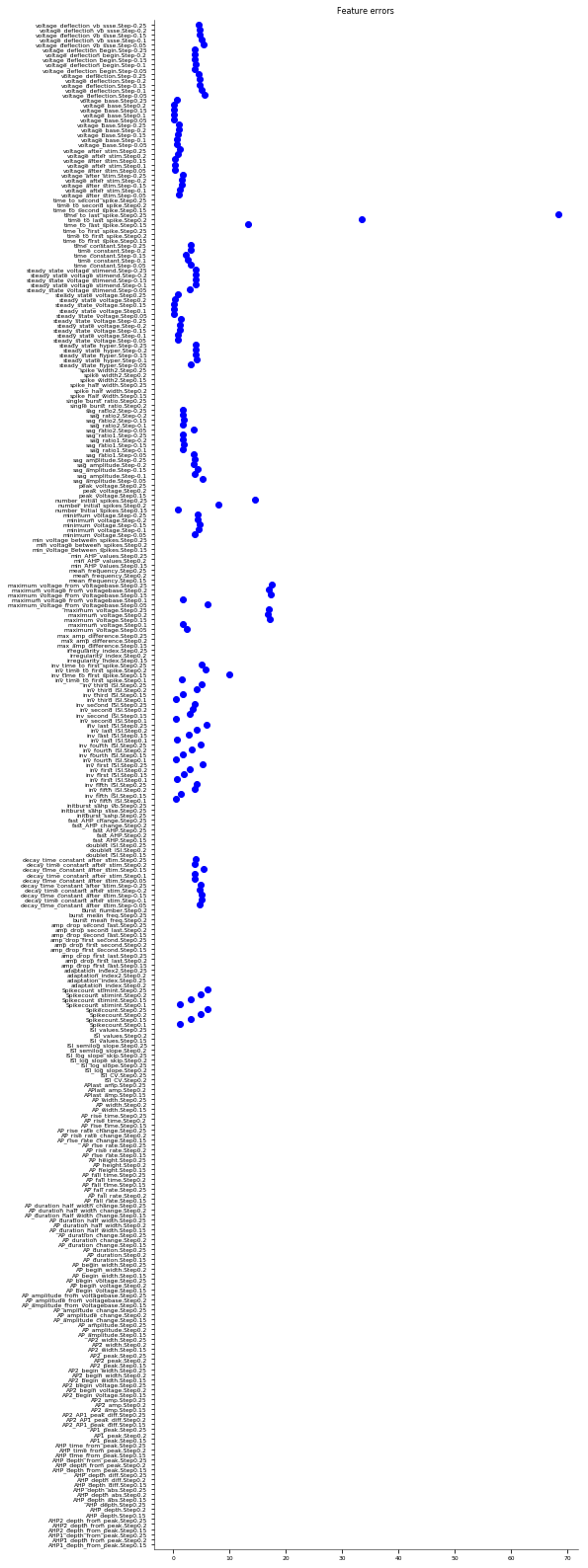
The figures are saved in the directory: ../published_models_validation_results/figs/somaticfeat_JMakara_data/Migliore_et_al_2011/
Features excluded (due to invalid values): AHP1_depth_from_peak.Step0.15, AHP1_depth_from_peak.Step0.2, AHP1_depth_from_peak.Step0.25, AHP2_depth_from_peak.Step0.15, AHP2_depth_from_peak.Step0.2, AHP2_depth_from_peak.Step0.25, AHP_depth.Step0.15, AHP_depth.Step0.2, AHP_depth.Step0.25, AHP_depth_abs.Step0.15, AHP_depth_abs.Step0.2, AHP_depth_abs.Step0.25, AHP_depth_diff.Step0.15, AHP_depth_diff.Step0.2, AHP_depth_diff.Step0.25, AHP_depth_from_peak.Step0.15, AHP_depth_from_peak.Step0.2, AHP_depth_from_peak.Step0.25, AHP_time_from_peak.Step0.15, AHP_time_from_peak.Step0.2, AHP_time_from_peak.Step0.25, AP1_peak.Step0.15, AP1_peak.Step0.2, AP1_peak.Step0.25, AP2_AP1_peak_diff.Step0.15, AP2_AP1_peak_diff.Step0.2, AP2_AP1_peak_diff.Step0.25, AP2_amp.Step0.15, AP2_amp.Step0.2, AP2_amp.Step0.25, AP2_begin_voltage.Step0.15, AP2_begin_voltage.Step0.2, AP2_begin_voltage.Step0.25, AP2_begin_width.Step0.15, AP2_begin_width.Step0.2, AP2_begin_width.Step0.25, AP2_peak.Step0.15, AP2_peak.Step0.2, AP2_peak.Step0.25, AP2_width.Step0.15, AP2_width.Step0.2, AP2_width.Step0.25, AP_amplitude.Step0.15, AP_amplitude.Step0.2, AP_amplitude.Step0.25, AP_amplitude_change.Step0.15, AP_amplitude_change.Step0.2, AP_amplitude_change.Step0.25, AP_amplitude_from_voltagebase.Step0.15, AP_amplitude_from_voltagebase.Step0.2, AP_amplitude_from_voltagebase.Step0.25, AP_begin_voltage.Step0.15, AP_begin_voltage.Step0.2, AP_begin_voltage.Step0.25, AP_begin_width.Step0.15, AP_begin_width.Step0.2, AP_begin_width.Step0.25, AP_duration.Step0.15, AP_duration.Step0.2, AP_duration.Step0.25, AP_duration_change.Step0.15, AP_duration_change.Step0.2, AP_duration_change.Step0.25, AP_duration_half_width.Step0.15, AP_duration_half_width.Step0.2, AP_duration_half_width.Step0.25, AP_duration_half_width_change.Step0.15, AP_duration_half_width_change.Step0.2, AP_duration_half_width_change.Step0.25, AP_fall_rate.Step0.15, AP_fall_rate.Step0.2, AP_fall_rate.Step0.25, AP_fall_time.Step0.15, AP_fall_time.Step0.2, AP_fall_time.Step0.25, AP_height.Step0.15, AP_height.Step0.2, AP_height.Step0.25, AP_rise_rate.Step0.15, AP_rise_rate.Step0.2, AP_rise_rate.Step0.25, AP_rise_rate_change.Step0.15, AP_rise_rate_change.Step0.2, AP_rise_rate_change.Step0.25, AP_rise_time.Step0.15, AP_rise_time.Step0.2, AP_rise_time.Step0.25, AP_width.Step0.15, AP_width.Step0.2, AP_width.Step0.25, APlast_amp.Step0.15, APlast_amp.Step0.2, APlast_amp.Step0.25, ISI_CV.Step0.2, ISI_CV.Step0.25, ISI_log_slope.Step0.2, ISI_log_slope.Step0.25, ISI_log_slope_skip.Step0.2, ISI_log_slope_skip.Step0.25, ISI_semilog_slope.Step0.2, ISI_semilog_slope.Step0.25, ISI_values.Step0.15, ISI_values.Step0.2, ISI_values.Step0.25, adaptation_index.Step0.2, adaptation_index.Step0.25, adaptation_index2.Step0.2, adaptation_index2.Step0.25, amp_drop_first_last.Step0.15, amp_drop_first_last.Step0.2, amp_drop_first_last.Step0.25, amp_drop_first_second.Step0.15, amp_drop_first_second.Step0.2, amp_drop_first_second.Step0.25, amp_drop_second_last.Step0.15, amp_drop_second_last.Step0.2, amp_drop_second_last.Step0.25, burst_mean_freq.Step0.2, burst_mean_freq.Step0.25, burst_number.Step0.2, doublet_ISI.Step0.15, doublet_ISI.Step0.2, doublet_ISI.Step0.25, fast_AHP.Step0.15, fast_AHP.Step0.2, fast_AHP.Step0.25, fast_AHP_change.Step0.2, fast_AHP_change.Step0.25, initburst_sahp.Step0.25, initburst_sahp_ssse.Step0.25, initburst_sahp_vb.Step0.25, irregularity_index.Step0.15, irregularity_index.Step0.2, irregularity_index.Step0.25, max_amp_difference.Step0.15, max_amp_difference.Step0.2, max_amp_difference.Step0.25, mean_frequency.Step0.15, mean_frequency.Step0.2, mean_frequency.Step0.25, min_AHP_values.Step0.15, min_AHP_values.Step0.2, min_AHP_values.Step0.25, min_voltage_between_spikes.Step0.15, min_voltage_between_spikes.Step0.2, min_voltage_between_spikes.Step0.25, peak_voltage.Step0.15, peak_voltage.Step0.2, peak_voltage.Step0.25, single_burst_ratio.Step0.2, single_burst_ratio.Step0.25, spike_half_width.Step0.15, spike_half_width.Step0.2, spike_half_width.Step0.25, spike_width2.Step0.15, spike_width2.Step0.2, spike_width2.Step0.25, time_to_first_spike.Step0.15, time_to_first_spike.Step0.2, time_to_first_spike.Step0.25, time_to_second_spike.Step0.15, time_to_second_spike.Step0.2, time_to_second_spike.Step0.25
Number of features succesfully evaluated: 142/305
Results are saved in the directory: ../published_models_validation_results/results/somaticfeat_JMakara_data/Migliore_et_al_2011/
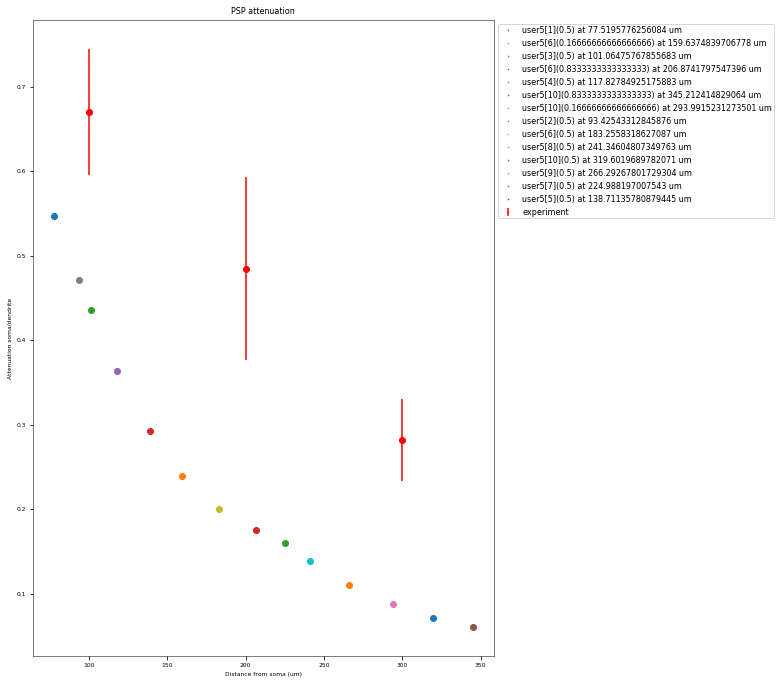
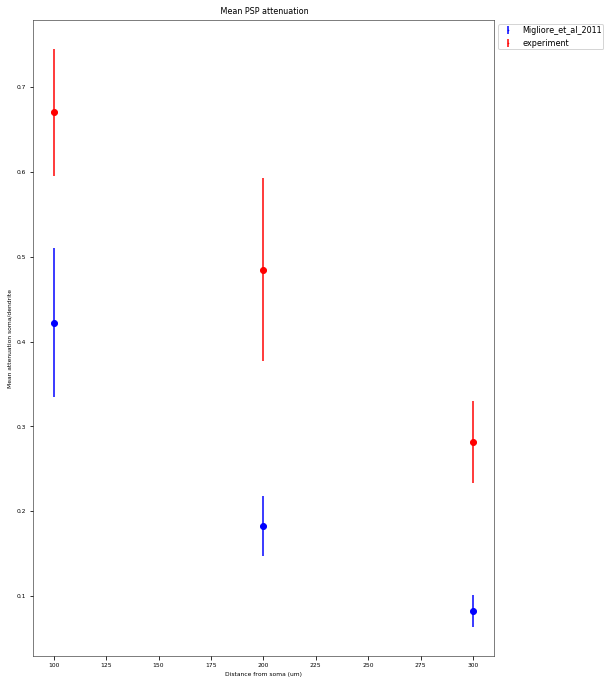
3.3.3. PSP Attenuatuion Test¶
The PSP Attenuation Test evaluates how much the post synaptic potential (using EPSC stimulus) attenuates from the main apical dendrite (different distances) to the soma. The soma/dendrite attenuation is compared to data from Magee & Cook 2000 (http://dx.doi.org/10.1038/78800).
[7]:
# Load target data
with open("../target_features/feat_PSP_attenuation_target_data.json", 'r') as f:
observation = json.load(f, object_pairs_hook=collections.OrderedDict)
IPython.display.HTML(json2html.convert(json = observation))
[7]:
| mean_attenuation_soma/dend_100_um | 0.67037891837 |
|---|---|
| std_attenuation_soma/dend_100_um | 0.0745536001493 |
| mean_attenuation_soma/dend_200_um | 0.485024302539 |
| std_attenuation_soma/dend_200_um | 0.108372269307 |
| mean_attenuation_soma/dend_300_um | 0.282118043034 |
| std_attenuation_soma/dend_300_um | 0.0482703550105 |
[8]:
# Load stimuli file
stim_file = pkg_resources.resource_filename("hippounit", "tests/stimuli/PSP_attenuation_stim/stim_PSP_attenuation_test.json")
with open(stim_file, 'r') as f:
config = json.load(f, object_pairs_hook=collections.OrderedDict)
# Instantiate test class
test = tests.PSPAttenuationTest(config=config, observation=observation, num_of_dend_locations = 15, force_run=False, show_plot=True, save_all = True, base_directory=base_directory, trunk_origin=trunk_origin)
# Number of parallel processes
test.npool = 10
try:
# Run the test
score = test.judge(model)
#Summarize and print the score achieved by the model on the test using SciUnit's summarize function
score.summarize()
except Exception as e:
print('Model: ' + model.name + ' could not be run')
print(e)
pass
Dendritic locations to be tested (with their actual distances): {('user5[10]', 0.5): 319.6019689782071, ('user5[6]', 0.5): 183.2558318627087, ('user5[1]', 0.5): 77.5195776256084, ('user5[3]', 0.5): 101.06475767855683, ('user5[10]', 0.16666666666666666): 293.9915231273501, ('user5[4]', 0.5): 117.82784925175883, ('user5[7]', 0.5): 224.988197007543, ('user5[2]', 0.5): 93.42543312845876, ('user5[6]', 0.8333333333333333): 206.8741797547396, ('user5[8]', 0.5): 241.34604807349763, ('user5[10]', 0.8333333333333333): 345.212414829064, ('user5[9]', 0.5): 266.29267801729304, ('user5[6]', 0.16666666666666666): 159.6374839706778, ('user5[5]', 0.5): 138.71135780879445}
input at: user5[10](0.5) with weight: 0.0
input at: user5[8](0.5) with weight: 0.0
input at: user5[10](0.16666666666666666) with weight: 0.0
input at: user5[3](0.5) with weight: 0.0
input at: user5[6](0.5) with weight: 0.0
input at: user5[10](0.8333333333333333) with weight: 0.0
input at: user5[7](0.5) with weight: 0.0
input at: user5[6](0.8333333333333333) with weight: 0.0
input at: user5[6](0.16666666666666666) with weight: 0.0
input at: user5[1](0.5) with weight: 0.0
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
input at: user5[9](0.5) with weight: 0.0
input at: user5[5](0.5) with weight: 0.0
input at: user5[2](0.5) with weight: 0.0
input at: user5[4](0.5) with weight: 0.0
Changed dt
Changed dt
Changed dt
Changed dt
input at: user5[6](0.5) with weight: 0.00046153846153846153
input at: user5[1](0.5) with weight: 0.00046153846153846153
input at: user5[3](0.5) with weight: 0.00046153846153846153
input at: user5[4](0.5) with weight: 0.00046153846153846153
input at: user5[6](0.8333333333333333) with weight: 0.00046153846153846153
input at: user5[10](0.8333333333333333) with weight: 0.00046153846153846153
input at: user5[10](0.16666666666666666) with weight: 0.00046153846153846153
input at: user5[6](0.16666666666666666) with weight: 0.00046153846153846153
input at: user5[2](0.5) with weight: 0.00046153846153846153
input at: user5[8](0.5) with weight: 0.00046153846153846153
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
input at: user5[10](0.5) with weight: 0.00046153846153846153
Changed dt
input at: user5[9](0.5) with weight: 0.00046153846153846153
input at: user5[7](0.5) with weight: 0.00046153846153846153
input at: user5[5](0.5) with weight: 0.00046153846153846153
Changed dt
Changed dt
Changed dt
The figures are saved in the directory: ../published_models_validation_results/figs/PSP_attenuation/Migliore_et_al_2011/
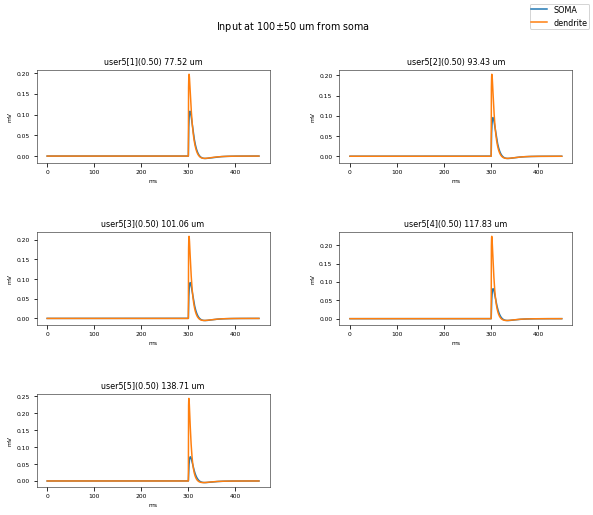
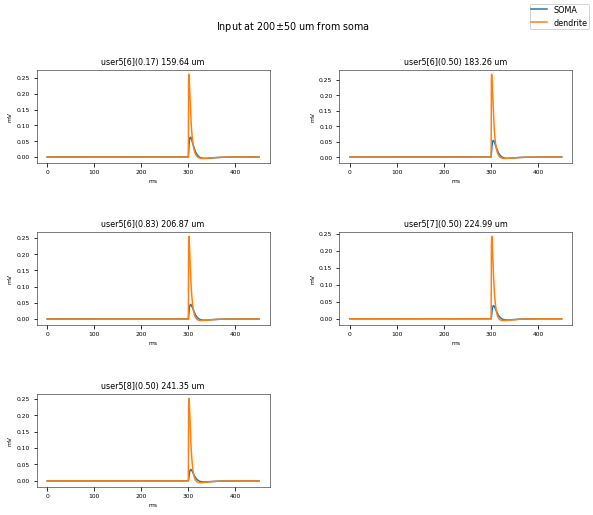
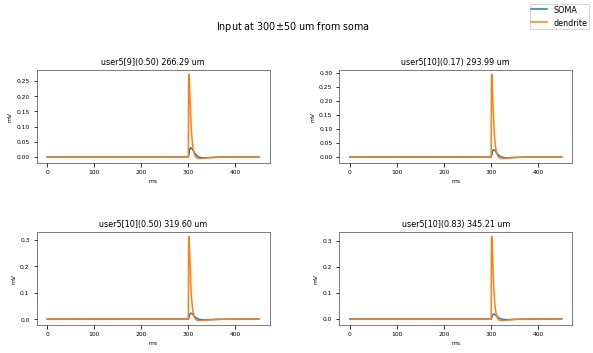
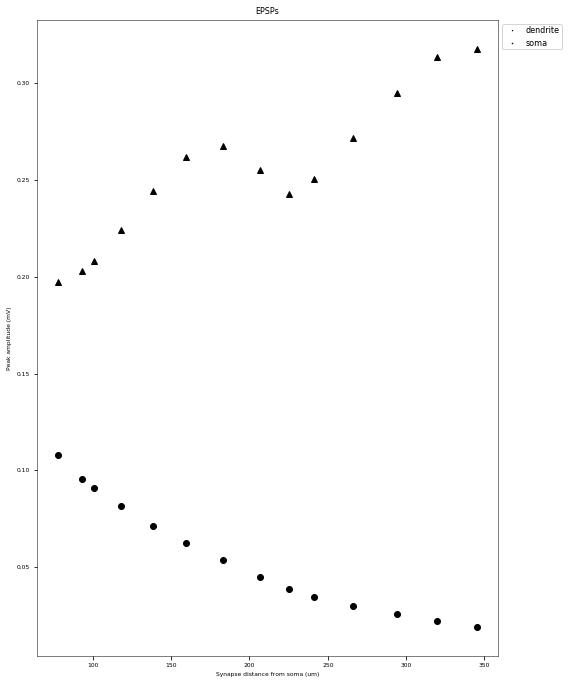

3.3.4. Back-Propagating AP Test¶
The Back-Propagating AP Test evaluates the mode and efficacy of back-propagating action potentials on the apical trunk in locations of different distances from the soma. The amplitude of the first and last AP of around 15 Hz train is compared to experimental data from Golding et al. 2001 (https://doi.org/10.1152/jn.2001.86.6.2998).
[9]:
# Load target data
with open('../target_features/feat_backpropagating_AP_target_data.json') as f:
observation = json.load(f, object_pairs_hook=collections.OrderedDict)
IPython.display.HTML(json2html.convert(json = observation))
[9]:
| mean_AP1_amp_at_50um | 66.6474010216 mV |
|---|---|
| std_AP1_amp_at_50um | 7.6800611053 mV |
| mean_AP1_amp_at_150um | 61.6405452338 mV |
| std_AP1_amp_at_150um | 8.84061901299 mV |
| mean_AP1_amp_at_250um | 57.1478276286 mV |
| std_AP1_amp_at_250um | 6.74288151869 mV |
| mean_AP1_amp_strong_propagating_at_350um | 52.5065653152 mV |
| std_AP1_amp_strong_propagating_at_350um | 5.82443856294 mV |
| mean_AP1_amp_weak_propagating_at_350um | 18.7832282514 mV |
| std_AP1_amp_weak_propagating_at_350um | 1.87203344953 mV |
| mean_APlast_amp_at_50um | 56.0027067193 mV |
| std_APlast_amp_at_50um | 6.66451614848 mV |
| mean_APlast_amp_at_150um | 41.6724147741 mV |
| std_APlast_amp_at_150um | 7.62175409041 mV |
| mean_APlast_amp_at_250um | 21.1507839635 mV |
| std_APlast_amp_at_250um | 2.39551093081 mV |
| mean_APlast_amp_at_350um | 9.81008709754 mV |
| std_APlast_amp_at_350um | 3.67378280696 mV |
[10]:
# Load stimuli file
stim_file = pkg_resources.resource_filename("hippounit", "tests/stimuli/bAP_stim/stim_bAP_test.json")
with open(stim_file, 'r') as f:
config = json.load(f, object_pairs_hook=collections.OrderedDict)
# Instantiate the test class
test = tests.BackpropagatingAPTest(config=config, observation=observation, force_run=False, force_run_FindCurrentStim=False, show_plot=True, save_all=True, base_directory=base_directory, trunk_origin=trunk_origin)
# Number of parallel processes
test.npool = 10
try:
# Run the test
score = test.judge(model)
#Summarize and print the score achieved by the model on the test using SciUnit's summarize function
score.summarize()
except Exception as e:
print('Model: ' + model.name + ' could not be run')
print(e)
pass
Dendritic locations to be tested (with their actual distances): {('user5[11]', 0.5): 368.0117721836399, ('user5[5]', 0.5): 138.71135780879445, ('user5[6]', 0.16666666666666666): 159.6374839706778, ('user5[8]', 0.5): 241.34604807349763, ('user5[10]', 0.8333333333333333): 345.212414829064, ('user5[9]', 0.5): 266.29267801729304, ('user5[0]', 0.5): 32.12825572041092}
Finding appropriate current step amplitude...
- running amplitude: 0.7000000000000001 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 0.2 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.0 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.5 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.30000000000000004 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.4 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.6000000000000001 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
- running amplitude: 0.9 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 0.1 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.8 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
Changed dt
- running amplitude: 1.0 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 0.4 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
The figures are saved in the directory: ../published_models_validation_results/figs/backpropagating_AP/Migliore_et_al_2011/
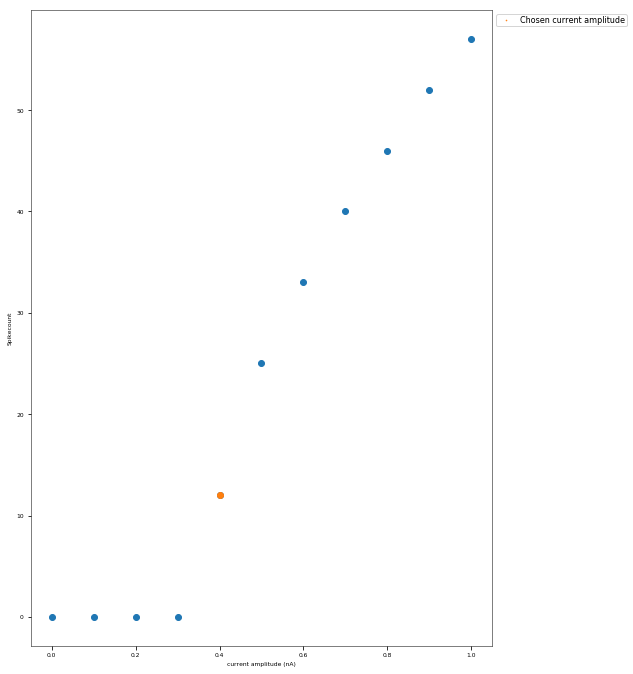
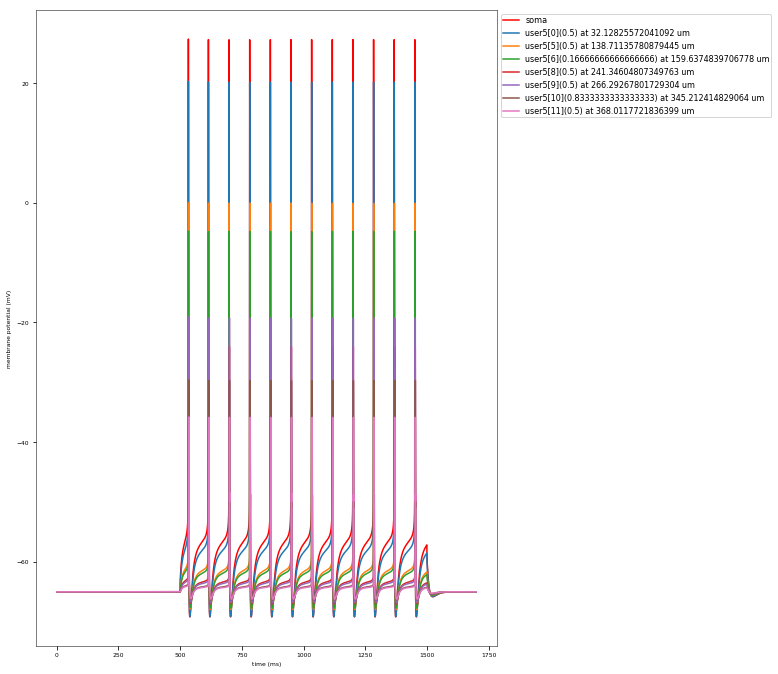
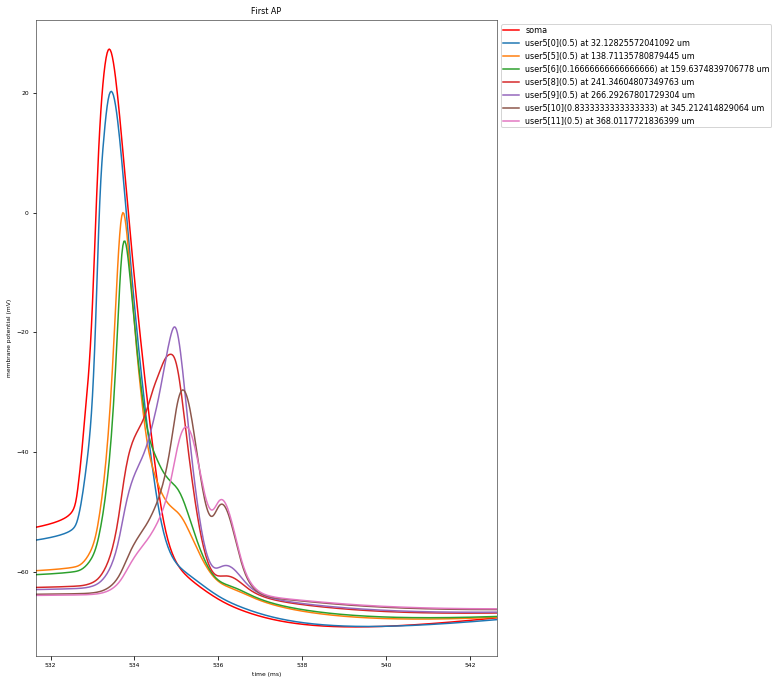
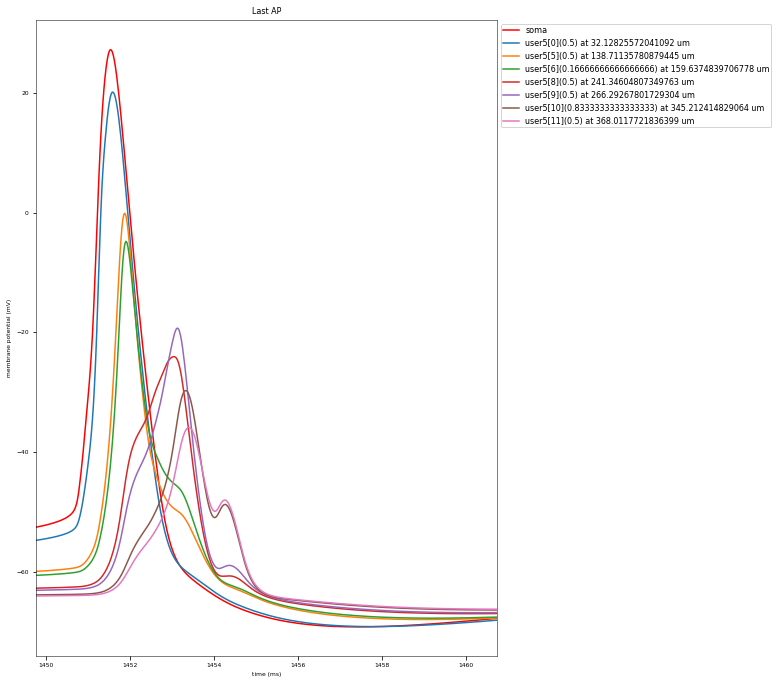
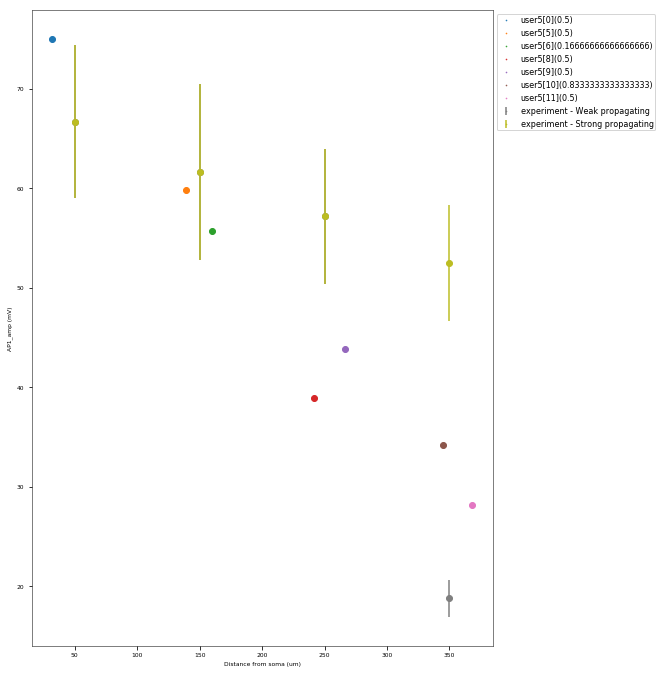
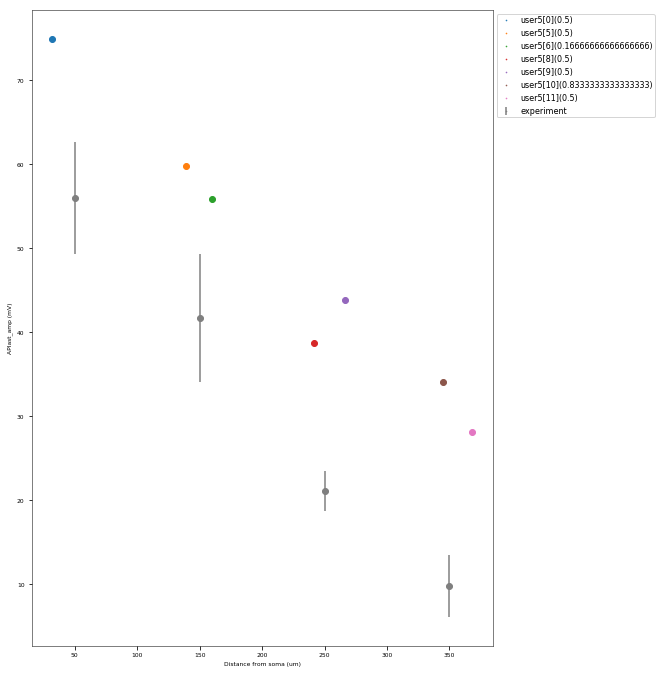
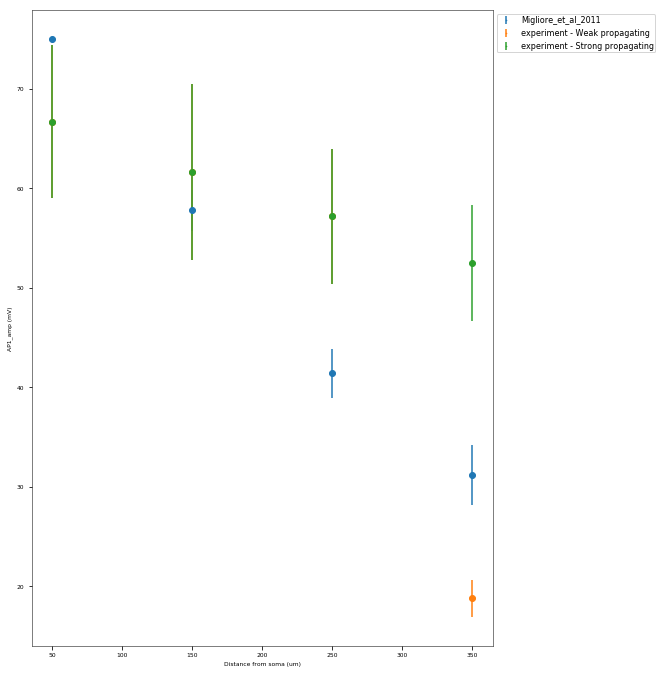
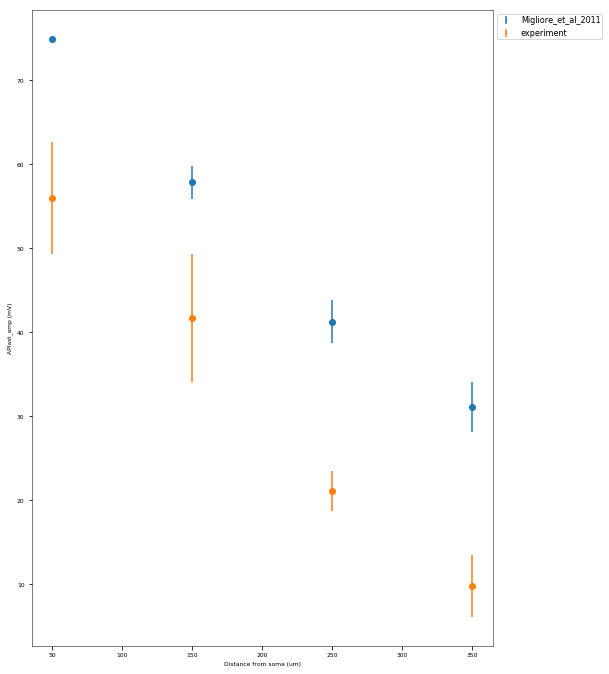
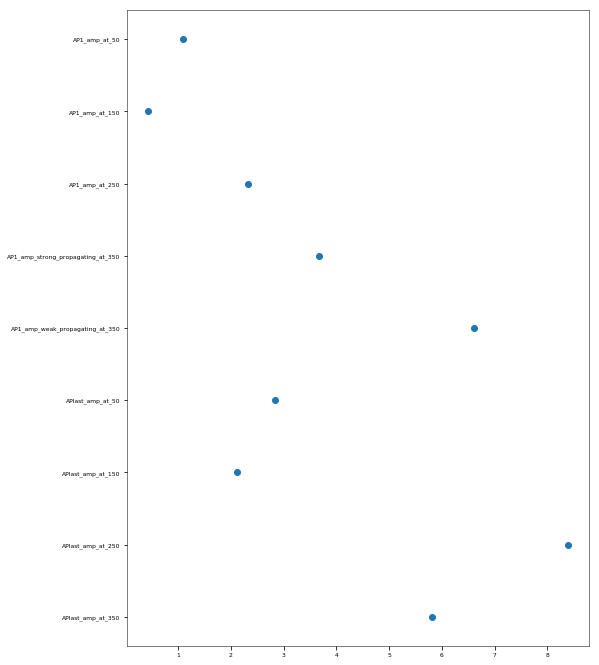
This is a rather STRONG propagating model
3.3.5. Depolarization-block Test¶
The Depolarization Block Test aims to determine whether the model enters depolarization block to prolonged, high intensity somatic current stimulus. It compares the current intensity to which the model fires the maximum number of action potentials, the current intensity before the model enters depolarization block (the two should be equal) and the equilibrium potential during depolarization block to the experimental data of Bianchi et al. 2012 (http://dx.doi.org/10.1007/s10827-012-0383-y).
[11]:
# Load target data
with open('../target_features/depol_block_target_data.json') as f:
observation = json.load(f, object_pairs_hook=collections.OrderedDict)
IPython.display.HTML(json2html.convert(json = observation))
[11]:
| mean_Ith | 0.6 nA |
|---|---|
| Ith_std | 0.3 nA |
| mean_Veq | -40.1 mV |
| Veq_std | 3.4 mV |
[12]:
# Instantiate the test class
test = tests.DepolarizationBlockTest(observation=observation, force_run=False, show_plot=True, save_all=True, base_directory=base_directory)
# Number of parallel processes
test.npool = 10
try:
# Run the test
score = test.judge(model)
#Summarize and print the score achieved by the model on the test using SciUnit's summarize function
score.summarize()
except Exception as e:
print('Model: ' + model.name + ' could not be run')
print(e)
pass
- running amplitude: 0.05 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.15000000000000002 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.0 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.35000000000000003 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.25 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.2 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.4 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.30000000000000004 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.1 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 0.45 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
- running amplitude: 0.5 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 0.55 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 0.6000000000000001 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 0.65 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 0.7000000000000001 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 0.75 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 0.8500000000000001 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 0.8 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 0.9 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 0.9500000000000001 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 1.0 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 1.05 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 1.1 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 1.1500000000000001 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 1.2000000000000002 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 1.25 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 1.35 on model: Migliore_et_al_2011 at: soma[0](0.5)
- running amplitude: 1.3 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
Changed dt
- running amplitude: 1.4000000000000001 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 1.4500000000000002 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 1.5 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 1.55 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
- running amplitude: 1.6 on model: Migliore_et_al_2011 at: soma[0](0.5)
Changed dt
The figures are saved in the directory: ../published_models_validation_results/figs/depol_block/Migliore_et_al_2011/
I_maxNumAP (the current intensity for which the model exhibited the maximum number of APs): 1.6 nA
I_below_depol_block (the current intensity before the model enters depolarization block): nan nA
Veq (the equilibrium value during the depolarization block): nan mV
Results are saved in the directory: ../published_models_validation_results/results/depol_block/Migliore_et_al_2011/
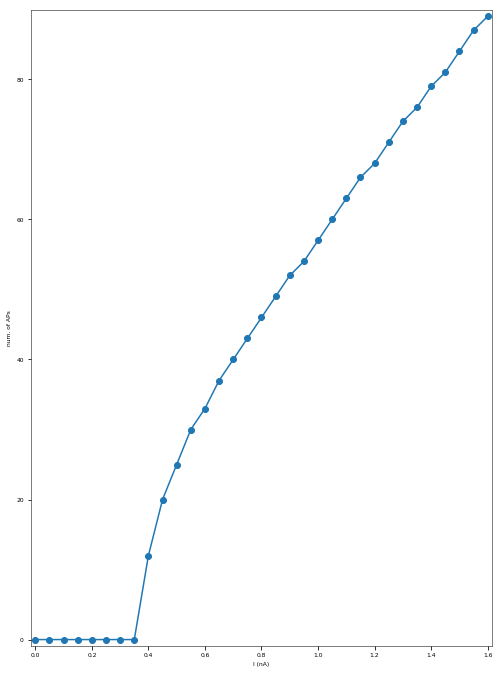
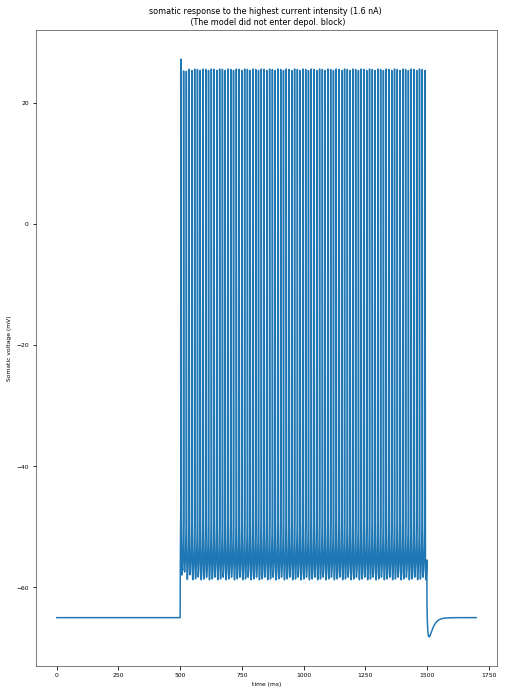
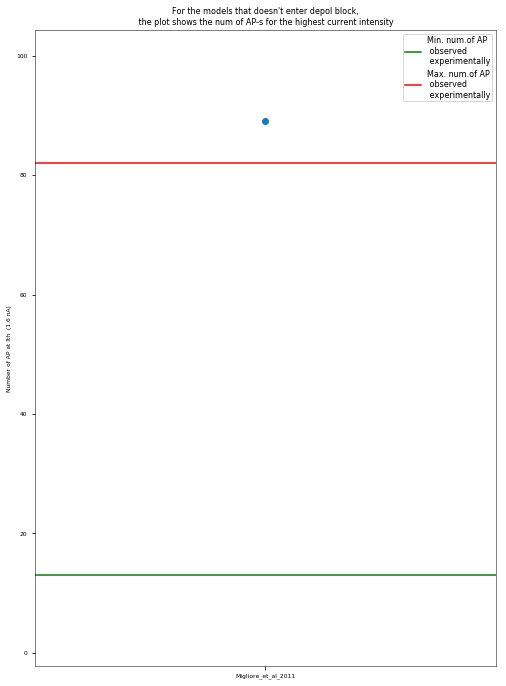

The model did not enter depolarization block.
3.3.6. Oblique Integration Test¶
Tests signal integration in oblique dendrites for increasing number of synchronous and asynchronous synaptic inputs. The experimental data is obtained from Losonczy and Magee 2006 (https://doi.org/10.1016/j.neuron.2006.03.016).
[13]:
# Load target data
with open('../target_features/oblique_target_data.json') as f:
observation = json.load(f, object_pairs_hook=collections.OrderedDict)
IPython.display.HTML(json2html.convert(json = observation))
[13]:
| mean_threshold | 3.4 mV |
|---|---|
| threshold_sem | 0.2 mV |
| exp_n | 92 |
| mean_prox_threshold | 4.5 mV |
| prox_threshold_sem | 0.5 mV |
| prox_n | 33 |
| mean_dist_threshold | 2.6 mV |
| dist_threshold_sem | 0.2 mV |
| dist_n | 44 |
| mean_nonlin_at_th | 142 |
| nonlin_at_th_sem | 9 |
| mean_nonlin_suprath | 129.0 |
| nonlin_suprath_sem | 6.0 |
| mean_peak_deriv | 2.6 V/s |
| peak_deriv_sem | 0.4 V/s |
| mean_amp_at_th | 4.828 mV |
| amp_at_th_sem | 0.28 mV |
| mean_time_to_peak | 13.5 ms |
| time_to_peak_sem | 0.2 ms |
| mean_async_nonlin | 104.0 |
| async_nonlin_sem | 8.0 |
| async_n | 23 |
[14]:
#setting synapse parameters
# If model.AMPA_name and/or model.NMDA_name is set here, the model's own receptor models (mod files) are used.
# If these are not set, HippoUnit's default synapse model is used. (AMPA: NEURON's Exp2Syn, NMDA: https://github.com/KaliLab/hippounit/blob/master/hippounit/tests/default_NMDAr/NMDA_CA1_pyr_SC.mod)
# Similarly to HippoUnit's default synapse, this model used the NEURON's Exp2Syn function as synaptic input,
# so here the same time constants are used as originally in the model.
# As the model doesn't have an NMDA receptor model, the default one of HippoUnit is used as the NMDA component of the synapse
model.AMPA_tau1 = 0.4
model.AMPA_tau2 = 1
# Instantiate the test class
test = tests.ObliqueIntegrationTest(observation = observation, save_all = True, force_run_synapse=False, force_run_bin_search=False, show_plot = True, base_directory = base_directory, trunk_origin=trunk_origin)
# Number of parallel processes
test.npool = 10
try:
# Run the test
score = test.judge(model)
#Summarize and print the score achieved by the model on the test using SciUnit's summarize function
score.summarize()
except Exception as e:
print('Model: ' + model.name + ' could not be run')
print(e)
pass
Dendrites and locations to be tested: [['apical_dendrite[17]', 0.30000000000000004, 'prox'], ['apical_dendrite[17]', 0.5, 'dist'], ['apical_dendrite[18]', 0.05555555555555555, 'prox'], ['apical_dendrite[18]', 0.2777777777777778, 'dist'], ['apical_dendrite[20]', 0.1, 'prox'], ['apical_dendrite[20]', 0.5, 'dist'], ['apical_dendrite[21]', 0.30000000000000004, 'prox'], ['apical_dendrite[21]', 0.5, 'dist'], ['apical_dendrite[22]', 0.30000000000000004, 'prox'], ['apical_dendrite[22]', 0.5, 'dist'], ['apical_dendrite[24]', 0.1, 'prox'], ['apical_dendrite[24]', 0.30000000000000004, 'dist'], ['apical_dendrite[25]', 0.21428571428571427, 'prox'], ['apical_dendrite[25]', 0.3571428571428571, 'dist'], ['apical_dendrite[28]', 0.1, 'prox'], ['apical_dendrite[28]', 0.30000000000000004, 'dist'], ['apical_dendrite[29]', 0.30000000000000004, 'prox'], ['apical_dendrite[29]', 0.5, 'dist'], ['apical_dendrite[30]', 0.1, 'prox'], ['apical_dendrite[30]', 0.5, 'dist']]
The built in Exp2Syn is used as the AMPA component. Tau1 = 0.4 ,Tau2 = 1 .
The default NMDA model of HippoUnit is used with Jahr, Stevens voltage dependence.
Adjusting synaptic weights on all the locations ...
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
The dendritic spike at dendrite apical_dendrite[18](0.05555555555555555) generated somatic AP - this location is not used in the test
The dendritic spike at dendrite apical_dendrite[28](0.1) generated somatic AP - this location is not used in the test
The dendritic spike at dendrite apical_dendrite[28](0.30000000000000004) generated somatic AP - this location is not used in the test
The dendritic spike at dendrite apical_dendrite[29](0.30000000000000004) generated somatic AP - this location is not used in the test
The dendritic spike at dendrite apical_dendrite[29](0.5) generated somatic AP - this location is not used in the test
The dendritic spike at dendrite apical_dendrite[30](0.1) generated somatic AP - this location is not used in the test
The dendritic spike at dendrite apical_dendrite[30](0.5) generated somatic AP - this location is not used in the test
- number of inputs: 0 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 1 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 2 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 4 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 7 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 8 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 3 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 5 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 6 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 9 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 0 dendrite: apical_dendrite[17] xloc: 0.5
- number of inputs: 1 dendrite: apical_dendrite[17] xloc: 0.5
- number of inputs: 2 dendrite: apical_dendrite[17] xloc: 0.5
Changed dt
Changed dt
Changed dt
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[17] xloc: 0.5
- number of inputs: 4 dendrite: apical_dendrite[17] xloc: 0.5
- number of inputs: 5 dendrite: apical_dendrite[17] xloc: 0.5
Changed dt
Changed dt
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[17] xloc: 0.5
- number of inputs: 7 dendrite: apical_dendrite[17] xloc: 0.5
- number of inputs: 8 dendrite: apical_dendrite[17] xloc: 0.5
Changed dt
Changed dt
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[17] xloc: 0.5
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[17] xloc: 0.5
- number of inputs: 0 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
- number of inputs: 1 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
Changed dt
Changed dt
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
- number of inputs: 4 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
- number of inputs: 5 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
- number of inputs: 6 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
Changed dt
Changed dt
Changed dt
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
- number of inputs: 0 dendrite: apical_dendrite[20] xloc: 0.1
- number of inputs: 1 dendrite: apical_dendrite[20] xloc: 0.1
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[20] xloc: 0.1
Changed dt
Changed dt
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[20] xloc: 0.1
- number of inputs: 4 dendrite: apical_dendrite[20] xloc: 0.1
Changed dt
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[20] xloc: 0.1
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[20] xloc: 0.1
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[20] xloc: 0.1
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[20] xloc: 0.1
- number of inputs: 9 dendrite: apical_dendrite[20] xloc: 0.1
- number of inputs: 10 dendrite: apical_dendrite[20] xloc: 0.1
- number of inputs: 0 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
Changed dt
Changed dt
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[20] xloc: 0.5
- number of inputs: 4 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[20] xloc: 0.5
- number of inputs: 9 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
- number of inputs: 0 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
- number of inputs: 1 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
- number of inputs: 10 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
Changed dt
- number of inputs: 0 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 0 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
- number of inputs: 5 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
- number of inputs: 8 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
- number of inputs: 9 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
Changed dt
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
- number of inputs: 0 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[22] xloc: 0.5
- number of inputs: 5 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[22] xloc: 0.5
- number of inputs: 10 dendrite: apical_dendrite[22] xloc: 0.5
- number of inputs: 0 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
Changed dt
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[24] xloc: 0.1
- number of inputs: 3 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[24] xloc: 0.1
- number of inputs: 9 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
- number of inputs: 0 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
- number of inputs: 0 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
- number of inputs: 6 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
- number of inputs: 0 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 5 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 7 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 2 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 3 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 0 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 8 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 4 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 6 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 9 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[17] xloc: 0.30000000000000004
- number of inputs: 0 dendrite: apical_dendrite[17] xloc: 0.5
- number of inputs: 1 dendrite: apical_dendrite[17] xloc: 0.5
Changed dt
Changed dt
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[17] xloc: 0.5
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[17] xloc: 0.5
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[17] xloc: 0.5
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[17] xloc: 0.5
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[17] xloc: 0.5
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[17] xloc: 0.5
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[17] xloc: 0.5
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[17] xloc: 0.5
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[17] xloc: 0.5
Changed dt
- number of inputs: 0 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
- number of inputs: 2 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
Changed dt
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
- number of inputs: 6 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
Changed dt
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
- number of inputs: 10 dendrite: apical_dendrite[18] xloc: 0.2777777777777778
Changed dt
Changed dt
- number of inputs: 0 dendrite: apical_dendrite[20] xloc: 0.1
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[20] xloc: 0.1
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[20] xloc: 0.1
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[20] xloc: 0.1
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[20] xloc: 0.1
- number of inputs: 5 dendrite: apical_dendrite[20] xloc: 0.1
Changed dt
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[20] xloc: 0.1
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[20] xloc: 0.1
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[20] xloc: 0.1
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[20] xloc: 0.1
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[20] xloc: 0.1
Changed dt
- number of inputs: 0 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[20] xloc: 0.5
Changed dt
- number of inputs: 0 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
- number of inputs: 2 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[21] xloc: 0.30000000000000004
Changed dt
- number of inputs: 0 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[21] xloc: 0.5
Changed dt
- number of inputs: 0 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
- number of inputs: 7 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[22] xloc: 0.30000000000000004
Changed dt
- number of inputs: 0 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[22] xloc: 0.5
Changed dt
- number of inputs: 0 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[24] xloc: 0.1
Changed dt
- number of inputs: 0 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
- number of inputs: 3 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
- number of inputs: 4 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
Changed dt
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[24] xloc: 0.30000000000000004
Changed dt
- number of inputs: 0 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
- number of inputs: 4 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[25] xloc: 0.21428571428571427
Changed dt
- number of inputs: 0 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 1 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 2 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 3 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 4 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 5 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 6 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 7 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 8 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 9 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
- number of inputs: 10 dendrite: apical_dendrite[25] xloc: 0.3571428571428571
Changed dt
The figures are saved in the directory: ../published_models_validation_results/figs/oblique_integration/Migliore_et_al_2011/
Results are saved in the directory: ../published_models_validation_results/results/oblique_integration/Migliore_et_al_2011/
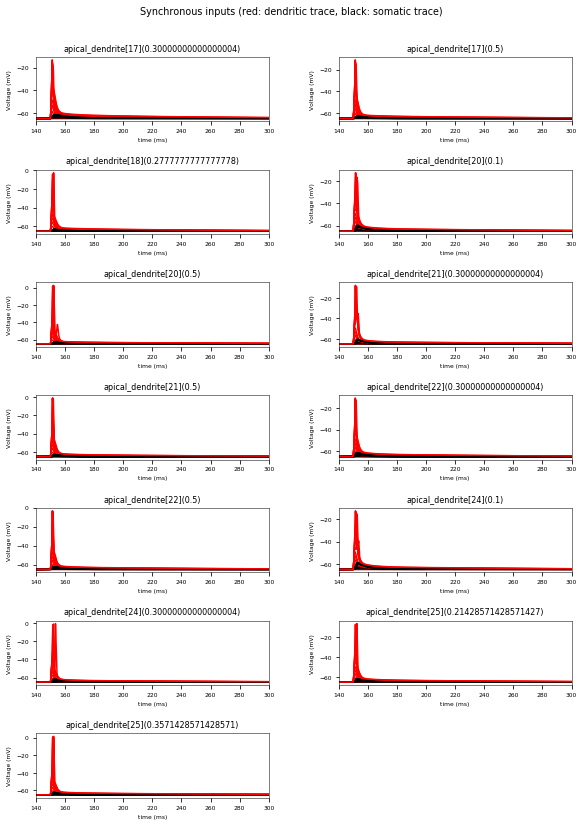
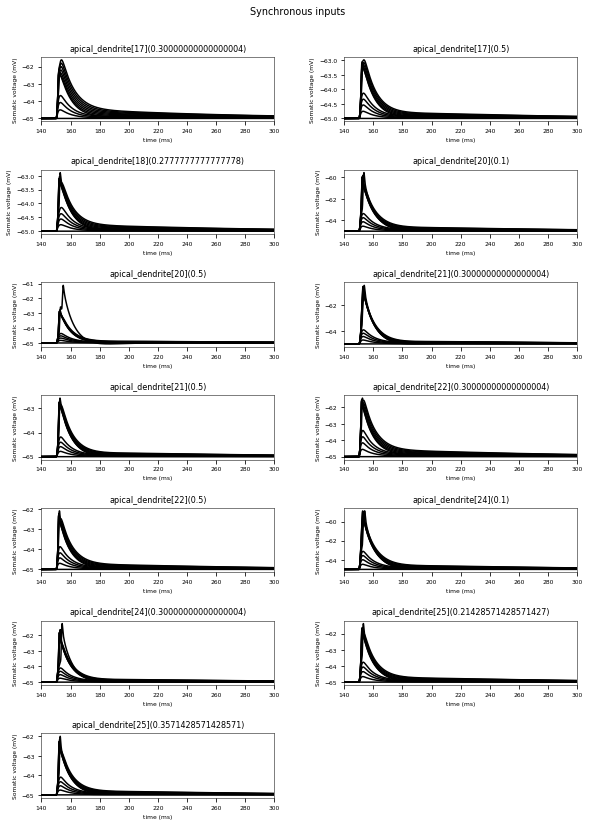
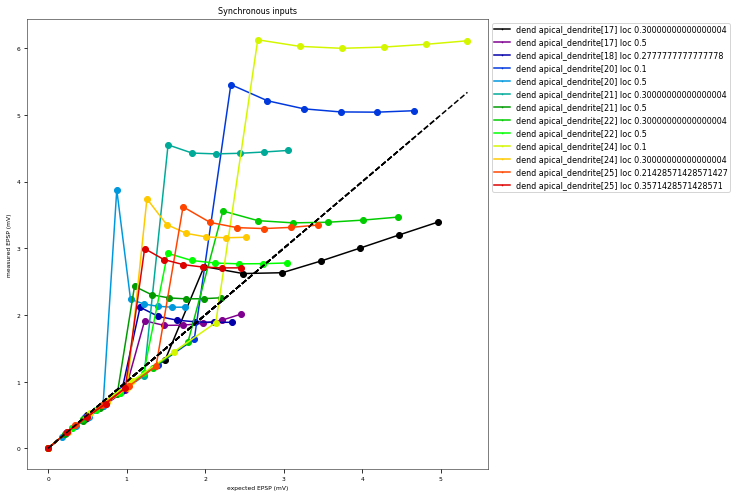
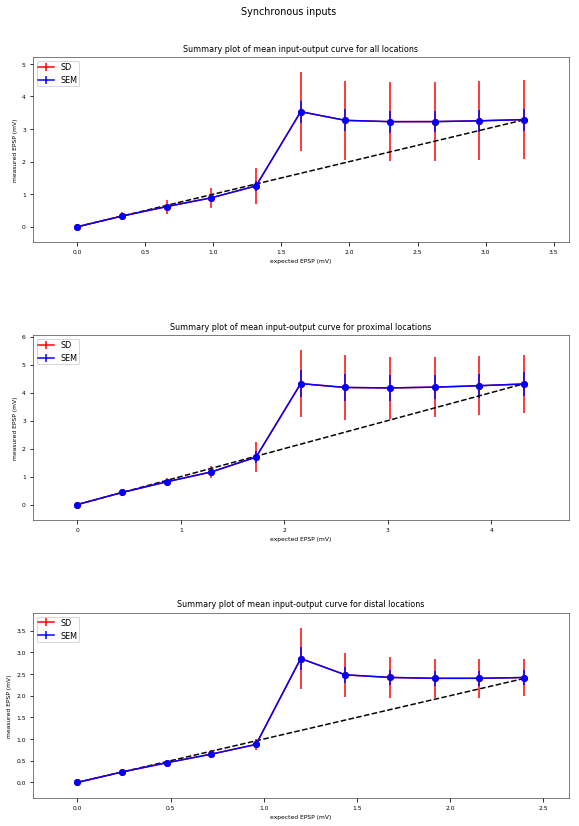
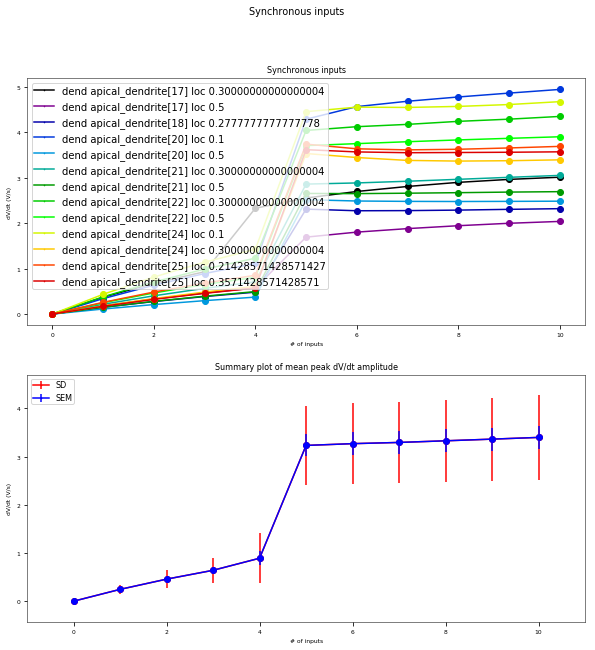
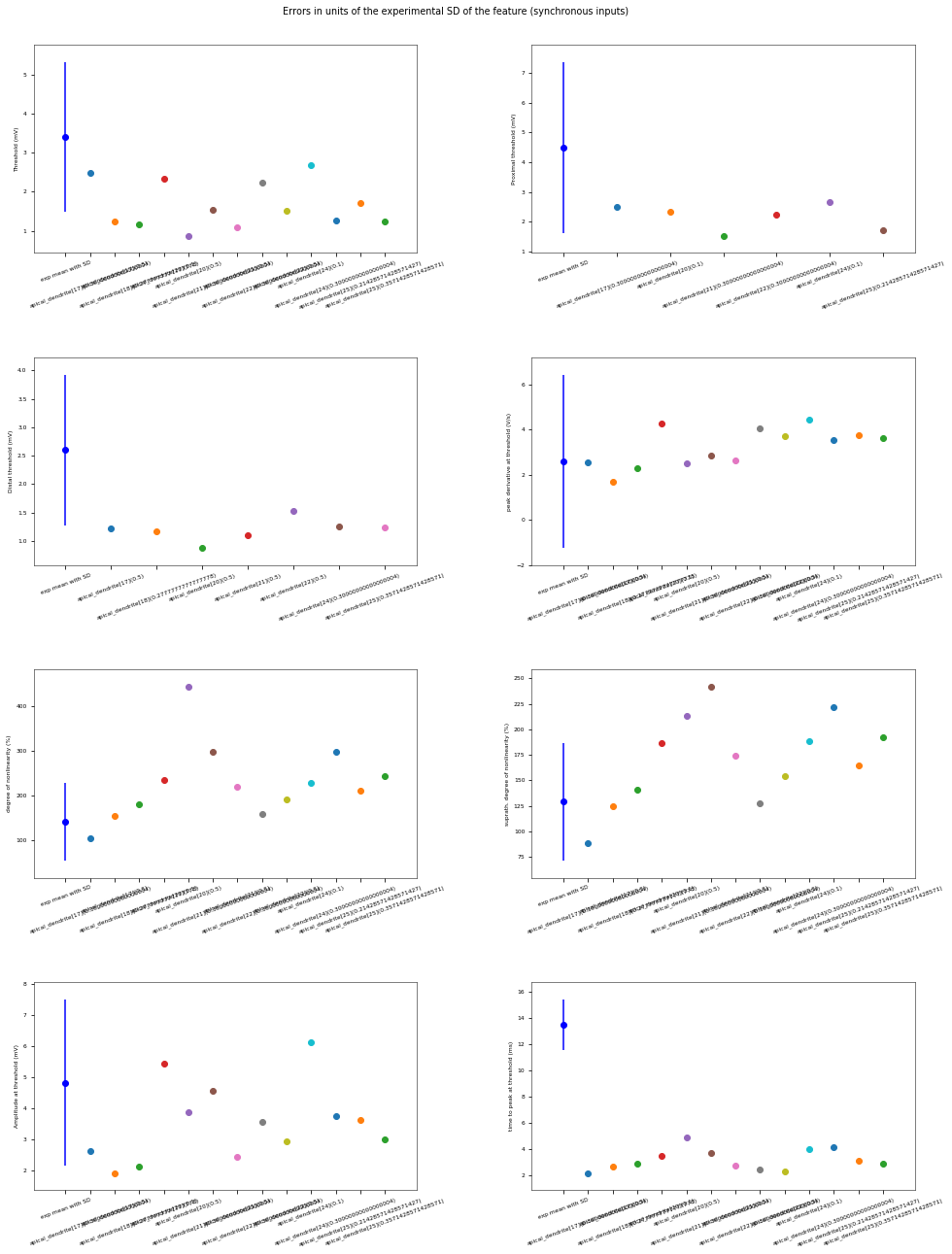
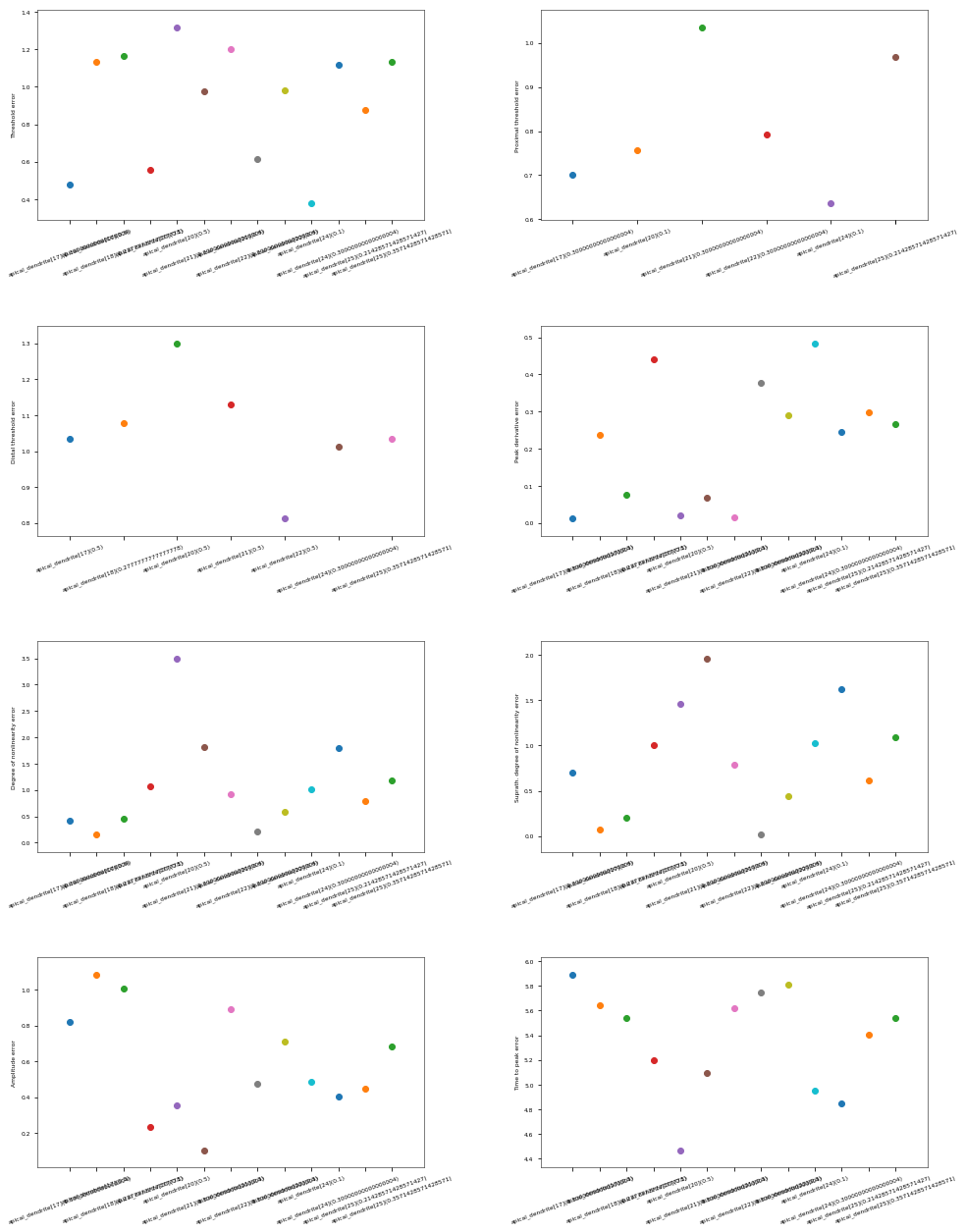
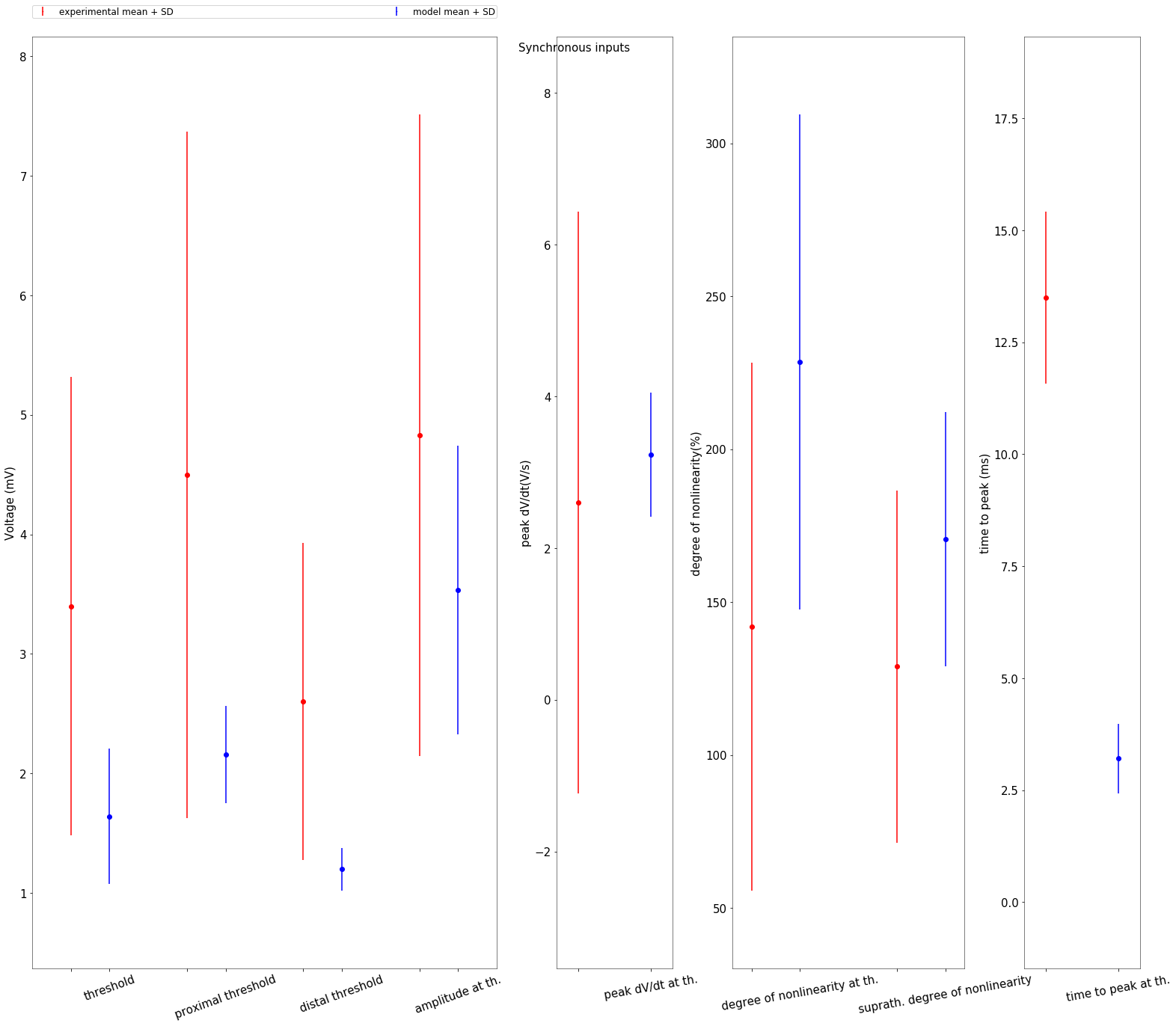
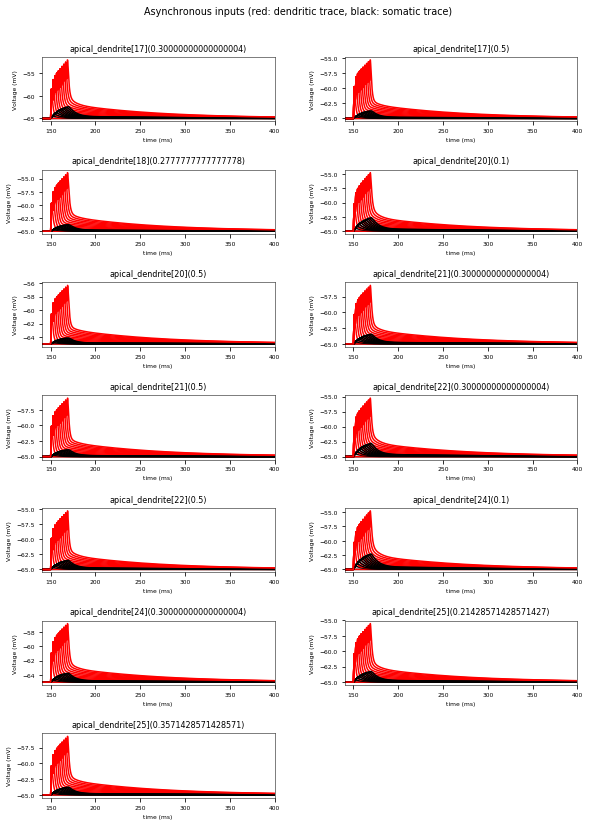
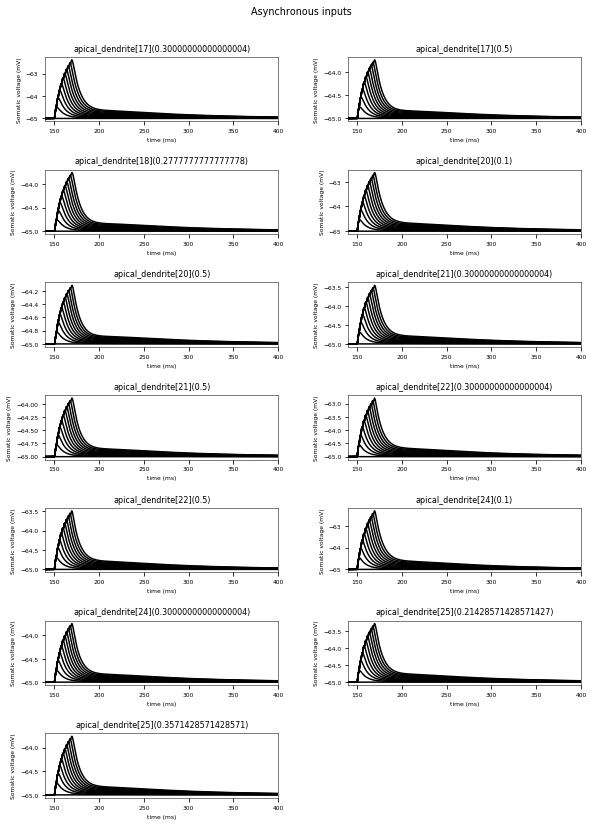
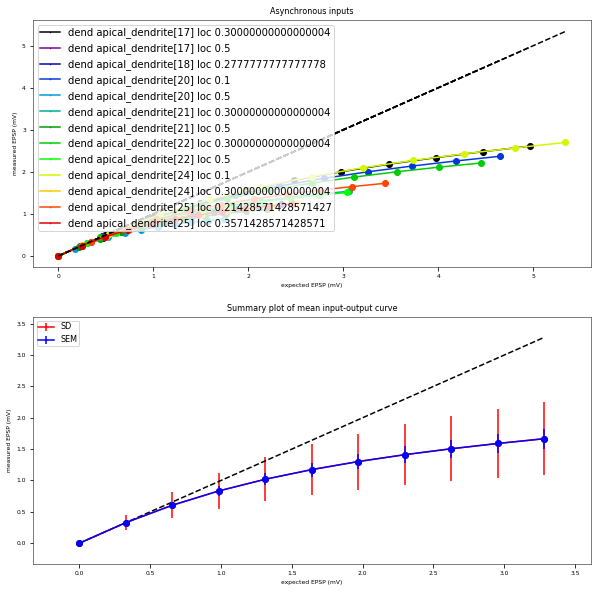
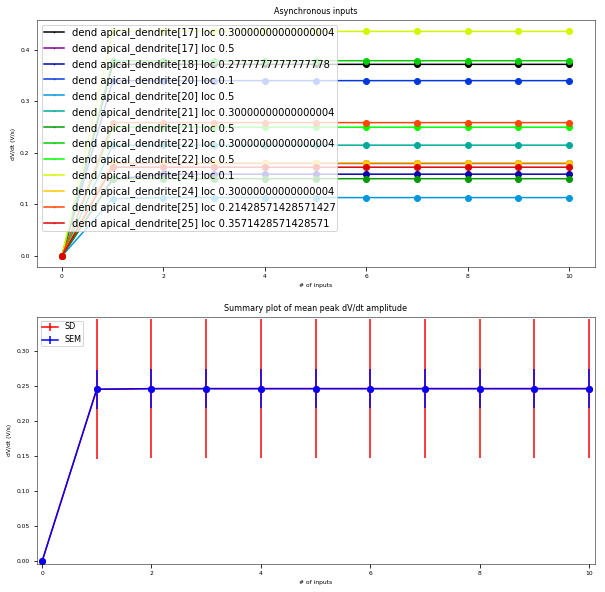
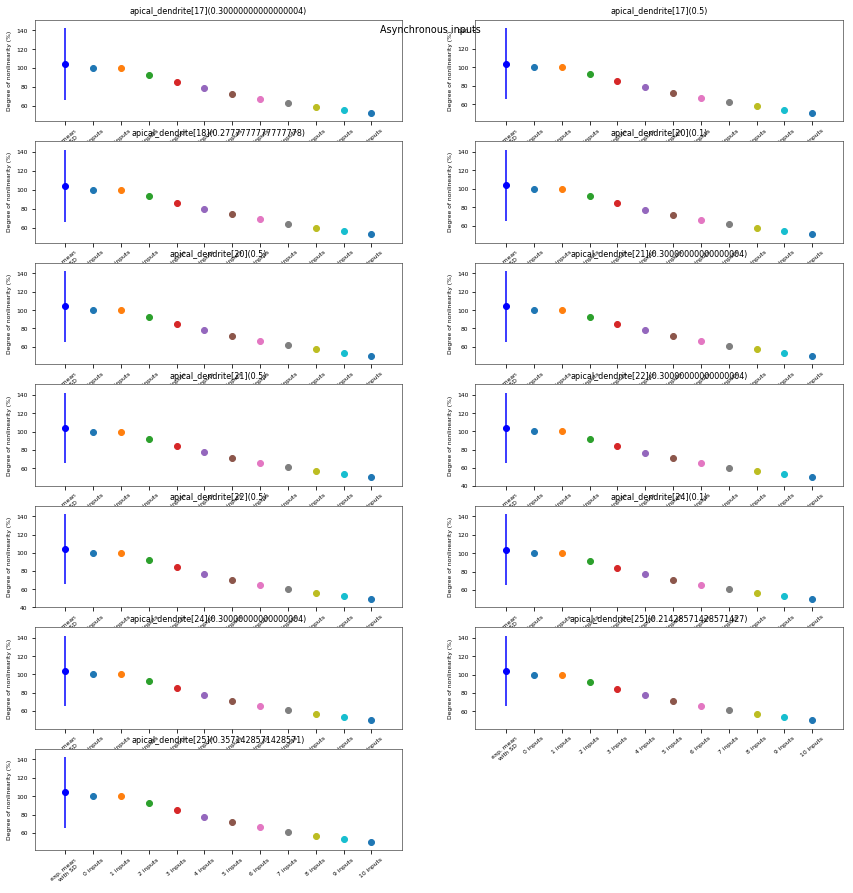
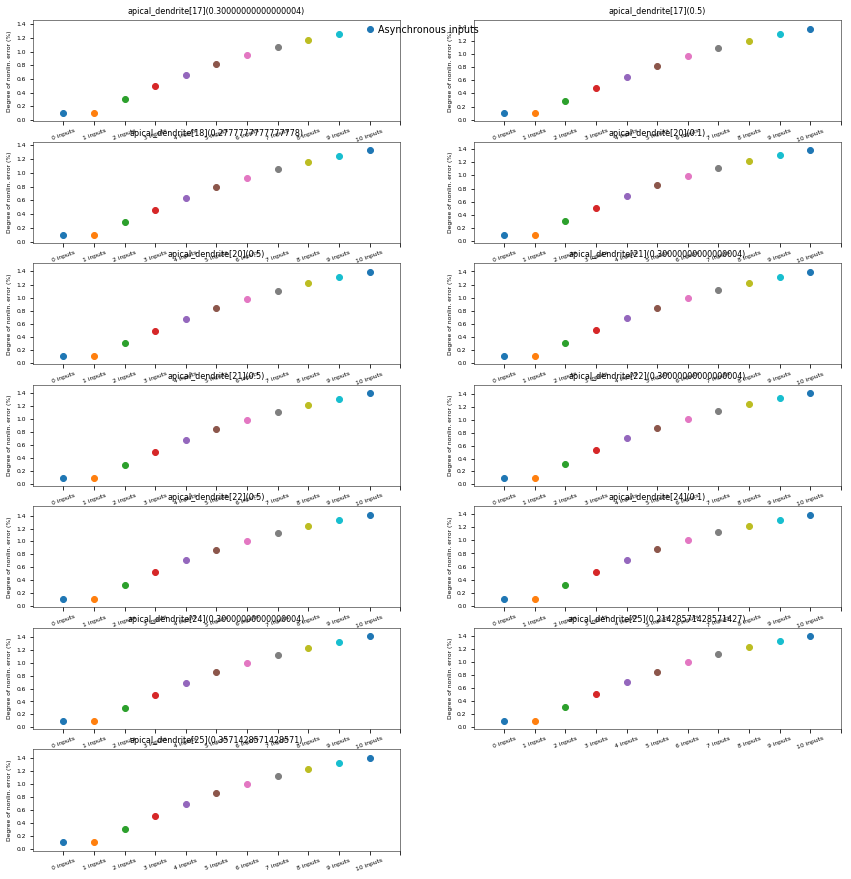
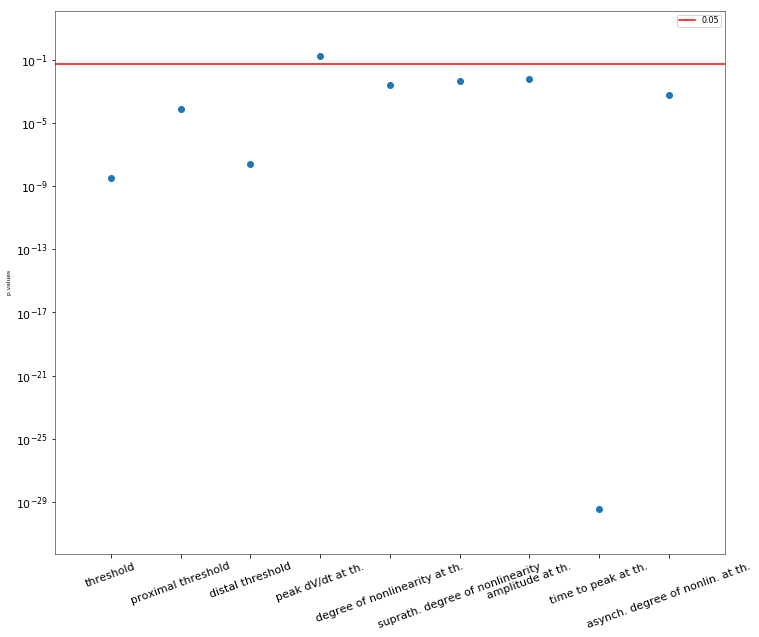
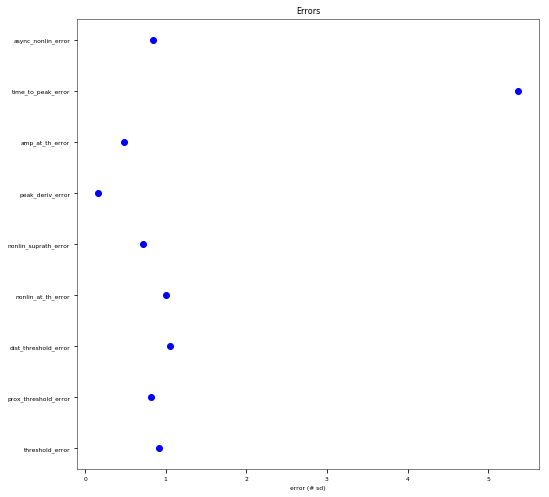
[15]:
%load_ext watermark
[16]:
%watermark -v -m -p sciunit,efel,numpy,quantities,scipy,matplotlib,neuron
CPython 3.5.2
IPython 5.6.0
sciunit 0.2.1.1
efel 3.0.58
numpy 1.16.4
quantities 0.12.1
scipy 1.3.0
matplotlib 3.0.3
neuron 7.6.2
compiler : GCC 5.4.0 20160609
system : Linux
release : 4.15.0-74-generic
machine : x86_64
processor : x86_64
CPU cores : 56
interpreter: 64bit